 |
PDBsum entry 1xvb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1xvb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 510 a.a.
510 a.a.
|
 |

|

|

|

|

|

|

|

 388 a.a.
388 a.a.
|
 |

|

|

|

|

|

|

|

 166 a.a.
166 a.a.
|
 |

|

|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Soluble methane monooxygenase hydroxylase: 6-bromohexanol soaked structure
|
|
Structure:
|
 |
Methane monooxygenase component a alpha chain. Chain: a, b. Synonym: methane hydroxylase. Methane monooxygenase component a beta chain. Chain: c, d. Synonym: methane hydroxylase. Methane monooxygenase component a gamma chain. Chain: e, f. Synonym: methane hydroxylase.
|
|
Source:
|
 |
Methylococcus capsulatus. Organism_taxid: 414. Organism_taxid: 414
|
|
Biol. unit:
|
 |
Hexamer (from
)
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.219
|
R-free:
|
0.251
|
|
|
Authors:
|
 |
M.H.Sazinsky,S.J.Lippard
|
|
Key ref:
|
 |
M.H.Sazinsky
and
S.J.Lippard
(2005).
Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): protein motion in the alpha-subunit.
J Am Chem Soc,
127,
5814-5825.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
27-Oct-04
|
Release date:
|
03-May-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P22869
(MEMA_METCA) -
Methane monooxygenase component A alpha chain from Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
527 a.a.
510 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.1.14.13.25
- methane monooxygenase (soluble).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
methane + NADH + O2 + H+ = methanol + NAD+ + H2O
|
|
2.
|
methane + NADPH + O2 + H+ = methanol + NADP+ + H2O
|
|
 |
 |
 |
 |
 |
methane
|
+
|
 NADH
NADH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
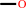 methanol
methanol
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
methane
|
+
|
 NADPH
NADPH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
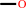 methanol
methanol
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Am Chem Soc
127:5814-5825
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Product bound structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): protein motion in the alpha-subunit.
|
|
M.H.Sazinsky,
S.J.Lippard.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The soluble methane monooxygenase hydroxylase (MMOH) alpha-subunit contains a
series of cavities that delineate the route of substrate entrance to and product
egress from the buried carboxylate-bridged diiron center. The presence of
discrete cavities is a major structural difference between MMOH, which can
hydroxylate methane, and toluene/o-xylene monooxygenase hydroxylase (ToMOH),
which cannot. To understand better the functions of the cavities and to
investigate how an enzyme designed for methane hydroxylation can also
accommodate larger substrates such as octane, methylcubane, and
trans-1-methyl-2-phenylcyclopropane, MMOH crystals were soaked with an
assortment of different alcohols and their X-ray structures were solved to
1.8-2.4 A resolution. The product analogues localize to cavities 1-3 and
delineate a path of product exit and/or substrate entrance from the active site
to the surface of the protein. The binding of the alcohols to a position
bridging the two iron atoms in cavity 1 extends and validates previous
crystallographic, spectroscopic, and computational work indicating this site to
be where substrates are hydroxylated and products form. The presence of these
alcohols induces perturbations in the amino acid side-chain gates linking pairs
of cavities, allowing for the formation of a channel similar to one observed in
ToMOH. Upon binding of 6-bromohexan-1-ol, the pi helix formed by residues
202-211 in helix E of the alpha-subunit is extended through residue 216,
changing the orientations of several amino acid residues in the active site
cavity. This remarkable secondary structure rearrangement in the four-helix
bundle has several mechanistic implications for substrate accommodation and the
function of the effector protein, MMOB.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.K.Boal,
J.A.Cotruvo,
J.Stubbe,
and
A.C.Rosenzweig
(2010).
Structural basis for activation of class Ib ribonucleotide reductase.
|
| |
Science,
329,
1526-1530.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Lasker,
A.Sali,
and
H.J.Wolfson
(2010).
Determining macromolecular assembly structures by molecular docking and fitting into an electron density map.
|
| |
Proteins,
78,
3205-3211.
|
 |
|
|
|
|
 |
R.B.Cooley,
B.L.Dubbels,
L.A.Sayavedra-Soto,
P.J.Bottomley,
and
D.J.Arp
(2009).
Kinetic characterization of the soluble butane monooxygenase from Thauera butanivorans, formerly 'Pseudomonas butanovora'.
|
| |
Microbiology,
155,
2086-2096.
|
 |
|
|
|
|
 |
L.J.Bailey,
J.G.McCoy,
G.N.Phillips,
and
B.G.Fox
(2008).
Structural consequences of effector protein complex formation in a diiron hydroxylase.
|
| |
Proc Natl Acad Sci U S A,
105,
19194-19198.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.J.Murray,
S.G.Naik,
D.O.Ortillo,
R.García-Serres,
J.K.Lee,
B.H.Huynh,
and
S.J.Lippard
(2007).
Characterization of the arene-oxidizing intermediate in ToMOH as a diiron(III) species.
|
| |
J Am Chem Soc,
129,
14500-14510.
|
 |
|
|
|
|
 |
K.H.Halsey,
L.A.Sayavedra-Soto,
P.J.Bottomley,
and
D.J.Arp
(2006).
Site-directed amino acid substitutions in the hydroxylase alpha subunit of butane monooxygenase from Pseudomonas butanovora: Implications for substrates knocking at the gate.
|
| |
J Bacteriol,
188,
4962-4969.
|
 |
|
|
|
|
 |
M.H.Sazinsky,
P.W.Dunten,
M.S.McCormick,
A.DiDonato,
and
S.J.Lippard
(2006).
X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase.
|
| |
Biochemistry,
45,
15392-15404.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.S.McCormick,
M.H.Sazinsky,
K.L.Condon,
and
S.J.Lippard
(2006).
X-ray crystal structures of manganese(II)-reconstituted and native toluene/o-xylene monooxygenase hydroxylase reveal rotamer shifts in conserved residues and an enhanced view of the protein interior.
|
| |
J Am Chem Soc,
128,
15108-15110.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Sobrado,
K.S.Lyle,
S.P.Kaul,
M.M.Turco,
I.Arabshahi,
A.Marwah,
and
B.G.Fox
(2006).
Identification of the binding region of the [2Fe-2S] ferredoxin in stearoyl-acyl carrier protein desaturase: insight into the catalytic complex and mechanism of action.
|
| |
Biochemistry,
45,
4848-4858.
|
 |
|
|
|
|
 |
G.Vardar,
Y.Tao,
J.Lee,
and
T.K.Wood
(2005).
Alanine 101 and alanine 110 of the alpha subunit of Pseudomonas stutzeri OX1 toluene-o-xylene monooxygenase influence the regiospecific oxidation of aromatics.
|
| |
Biotechnol Bioeng,
92,
652-658.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
| |

