 |
PDBsum entry 1xpy

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Isomerase
|
 |
|
Title:
|
 |
Structural basis for catalytic racemization and substrate specificity of an n-acylamino acid racemase homologue from deinococcus radiodurans
|
|
Structure:
|
 |
N-acylamino acid racemase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Deinococcus radiodurans. Organism_taxid: 1299. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Octamer (from
)
Octamer (from
)
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.174
|
R-free:
|
0.227
|
|
|
Authors:
|
 |
W.-C.Wang,W.-C.Chiu,S.-K.Hsu,C.-L.Wu,C.-Y.Chen,J.-S.Liu,W.-H.Hsu
|
Key ref:

|
 |
W.C.Wang
et al.
(2004).
Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans.
J Mol Biol,
342,
155-169.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
10-Oct-04
|
Release date:
|
26-Oct-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9RYA6
(Q9RYA6_DEIRA) -
N-succinylamino acid racemase from Deinococcus radiodurans (strain ATCC 13939 / DSM 20539 / JCM 16871 / CCUG 27074 / LMG 4051 / NBRC 15346 / NCIMB 9279 / VKM B-1422 / R1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
375 a.a.
360 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 5 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.113
- o-succinylbenzoate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1R,6R)-6-hydroxy-2-succinyl-cyclohexa-2,4-diene-1-carboxylate = 2-succinylbenzoate + H2O
|
 |
 |
 |
 |
 |
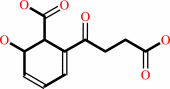 (1R,6R)-6-hydroxy-2-succinyl-cyclohexa-2,4-diene-1-carboxylate
(1R,6R)-6-hydroxy-2-succinyl-cyclohexa-2,4-diene-1-carboxylate
|
=
|
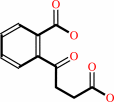 2-succinylbenzoate
2-succinylbenzoate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mn(2+) or Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
342:155-169
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans.
|
|
W.C.Wang,
W.C.Chiu,
S.K.Hsu,
C.L.Wu,
C.Y.Chen,
J.S.Liu,
W.H.Hsu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
N-acylamino acid racemase (NAAAR) catalyzes the racemization of N-acylamino
acids and can be used in concert with an aminoacylase to produce enantiopure
alpha-amino acids, a process that has potential industrial applications. Here we
have cloned and characterized an NAAAR homologue from a radiation-resistant
ancient bacterium, Deinococcus radiodurans. The expressed NAAAR racemized
various substrates at an optimal temperature of 60 degrees C and had Km values
of 24.8 mM and 12.3 mM for N-acetyl-D-methionine and N-acetyl-L-methionine,
respectively. The crystal structure of NAAAR was solved to 1.3 A resolution
using multiwavelength anomalous dispersion (MAD) methods. The structure consists
of a homooctamer in which each subunit has an architecture characteristic of
enolases with a capping domain and a (beta/alpha)7 beta barrel domain. The
NAAAR.Mg2+ and NAAAR.N-acetyl-L-glutamine.Mg2+ structures were also determined,
allowing us to define the Lys170-Asp195-Glu220-Asp245-Lys269 framework for
catalyzing 1,1-proton exchange of N-acylamino acids. Four subsites enclosing the
substrate are identified: catalytic site, metal-binding site, side-chain-binding
region, and a flexible lid region. The high conservation of catalytic and
metal-binding sites in different enolases reflects the essentiality of a common
catalytic platform, allowing these enzymes to robustly abstract alpha-protons of
various carboxylate substrates efficiently. The other subsites involved in
substrate recognition are less conserved, suggesting that divergent evolution
has led to functionally distinct enzymes.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. The binding pocket in the
NAAAR·NAQ·Mg2+ ternary complex. (a) Stereoview of
the active site of the NAAAR·NAQ·Mg2+ complex. NAQ
(dark green) is shown as a ball-and-stick model with carbon
atoms colored white. Five loops enclosing NAQ are green.
Residues in C sub-site (red), M sub-site (blue), S sub-site
(purple), and L region (yellow) are shown as stick models. The
oxygen and nitrogen atoms are red and blue, respectively. The
magnesium ion is shown in green. (b) Schematic representation of
interactions between NAQ and NAAAR. The color representation of
the four sub-sites is as in (a). The hydrogen-bonding
interactions are shown as broken lines.
|
 |
Figure 8.
Figure 8. Comparison of binding pockets in enolases. (a)
Superposition of binding pockets in the apo structures of NAAAR
(green), MLE (brown), YkfB (pink), and YcjG (cyan). The S and L
regions are shown as C^a traces and conserved carboxylate
residues and two catalytic Lys residues corresponding to Lys170,
Asp195, Glu220, Asp245, and Lys269 in NAAAR are shown as stick
models. The oxygen, nitrogen, and sulfur atoms are red, blue,
and yellow, respectively. (b) The binding pockets of NAAAR
(green), MAL (cyan), enolase (brown), and MR (pink) were
superimposed. The S and L (L1 and L2) sites are shown as C^a
traces and conserved carboxylate residues and catalytic residues
corresponding to Lys170, Asp195, Glu220, Asp245, and Lys269 in
NAAAR are shown as stick models. The bound ligand is shown as
thin sticks.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2004,
342,
155-169)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
I.C.Chen,
W.D.Lin,
S.K.Hsu,
V.Thiruvengadam,
and
W.H.Hsu
(2009).
Isolation and characterization of a novel lysine racemase from a soil metagenomic library.
|
| |
Appl Environ Microbiol,
75,
5161-5166.
|
 |
|
|
|
|
 |
J.Pozo-Dengra,
S.Martínez-Rodríguez,
L.M.Contreras,
J.Prieto,
M.Andújar-Sánchez,
J.M.Clemente-Jiménez,
F.J.Las Heras-Vázquez,
F.Rodríguez-Vico,
and
J.L.Neira
(2009).
Structure and conformational stability of a tetrameric thermostable N-succinylamino acid racemase.
|
| |
Biopolymers,
91,
757-772.
|
 |
|
|
|
|
 |
M.Hayashida,
S.H.Kim,
K.Takeda,
T.Hisano,
and
K.Miki
(2008).
Crystal structure of N-acylamino acid racemase from Thermus thermophilus HB8.
|
| |
Proteins,
71,
519-523.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.K.Hsu,
H.H.Lo,
C.H.Kao,
D.S.Lee,
and
W.H.Hsu
(2006).
Enantioselective synthesis of L-homophenylalanine by whole cells of recombinant Escherichia coli expressing L-aminoacylase and N-acylamino acid racemase genes from Deinococcus radiodurans BCRC12827.
|
| |
Biotechnol Prog,
22,
1578-1584.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

