 |
PDBsum entry 1xhl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1xhl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of putative tropinone reductase-ii from caenorhabditis elegans with cofactor and substrate
|
|
Structure:
|
 |
Short-chain dehydrogenase/reductase family member (5l265), putative tropinone reductase-ii. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Caenorhabditis elegans. Organism_taxid: 6239. Gene: f25d1.5. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Tetramer (from PDB file)
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.191
|
R-free:
|
0.215
|
|
|
Authors:
|
 |
N.Schormann,E.Karpova,J.Zhou,Y.Zhang,J.Symersky,R.Bunzel,W.-Y.Huang, A.Arabshahi,S.Qiu,C.-H.Luan,R.Gray,M.Carson,J.Tsao,M.Luo,D.Johnson, S.Lu,G.Lin,D.Luo,Z.Cao,S.Li,A.Mckinstry,Q.Shang,Y.-J.Chen,T.Bray, L.Nagy,L.Delucas,Southeast Collaboratory For Structural Genomics (Secsg)
|
|
Key ref:
|
 |
N.Schormann
et al.
Crystal structure of putative tropinone reductase-Ii from caenorhabditis elegans with cofactor and substrate.
To be published,
.
|
 |
|
Date:
|
 |
|
20-Sep-04
|
Release date:
|
28-Sep-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q19774
(Q19774_CAEEL) -
3-oxoacyl-[acyl-carrier-protein] reductase from Caenorhabditis elegans
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
277 a.a.
274 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.236
- tropinone reductase Ii.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
pseudotropine + NADP+ = tropinone + NADPH + H+
|
 |
 |
 |
 |
 |
pseudotropine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
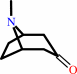 tropinone
tropinone
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

