 |
PDBsum entry 1xes

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of stilbene synthase from pinus sylvestris
|
|
Structure:
|
 |
Dihydropinosylvin synthase. Chain: a, b, c, d. Synonym: stilbene synthase, sts, pinosylvin-forming stilbene synthase. Engineered: yes
|
|
Source:
|
 |
Pinus sylvestris. Scots pine. Organism_taxid: 3349. Gene: sts. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.202
|
R-free:
|
0.225
|
|
|
Authors:
|
 |
S.H.Ng,D.Chirgadze,D.Spiteller,T.L.Li,J.B.Spencer,T.L.Blundell
|
|
Key ref:
|
 |
S.H.Ng
et al.
Crystal structure of stilbene synthase from pinus sylvestris.
To be published,
.
|
 |
|
Date:
|
 |
|
12-Sep-04
|
Release date:
|
11-Apr-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q02323
(DPSS_PINSY) -
Pinosylvin synthase from Pinus sylvestris
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
393 a.a.
387 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.146
- pinosylvin synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Chalcone and Stilbene Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(E)-cinnamoyl-CoA + 3 malonyl-CoA + 3 H+ = (E)-pinosylvin + 4 CO2 + 4 CoA
|
 |
 |
 |
 |
 |
 (E)-cinnamoyl-CoA
(E)-cinnamoyl-CoA
|
+
|
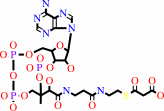 3
×
malonyl-CoA
3
×
malonyl-CoA
|
+
|
3
×
H(+)
|
=
|
(E)-pinosylvin
|
+
|
 4
×
CO2
4
×
CO2
|
+
|
 4
×
CoA
4
×
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

