 |
PDBsum entry 1xaa

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1xaa
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.85
- 3-isopropylmalate dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-isopropylmalate + NAD+ = 4-methyl-2-oxopentanoate + CO2 + NADH
|
 |
 |
 |
 |
 |
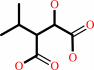 (2R,3S)-3-isopropylmalate
(2R,3S)-3-isopropylmalate
|
+
|
 NAD(+)
NAD(+)
|
=
|
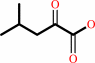 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 CO2
CO2
|
+
|
 NADH
NADH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
52:623-630
(1996)
|
|
PubMed id:
|
|
|
|

|
| |
|
Cryocrystallography of 3-Isopropylmalate dehydrogenase from Thermus thermophilus and its chimeric enzyme.
|
|
C.Nagata,
H.Moriyama,
N.Tanaka,
M.Nakasako,
M.Yamamoto,
T.Ueki,
T.Oshima.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of thermostable enzyme, 3-isopropylmalate dehydrogenase
of Thermus thermophilus (10T) and a chimeric enzyme between T. thermophilus and
Bacillus subtilus with one point mutation (cS82R), were determined at both 100
and 150 K. At the cryogenic condition, the volume of the unit cell decreased by
5% as a result of a contraction in the solvent region. Although the overall
structures of both enzymes at low temperature were the same as that of 10T at
room temperature, interactions between two domains and between two subunits in a
functional dimer of cS82R were significantly altered. The decrease in the
average temperature factor of 10T at low temperature and no significant decrease
for cS82R suggested that the structure of the engineered enzyme (cS82R) may have
many conformational substates even at low temperature, while the native enzyme
(10T) at low temperature has a more definite conformation than that at room
temperature. The location of water molecules around the enzyme molecule and the
calculation of the radii of gyration suggested that cS82R had a weaker hydration
than 10T.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 6.
Fig. 6. The temperature factors of the amino-acid residues. The solid
lines indicate those at 100 K and the dotted lines indicate those at
293 K. (a) 10T. (b) cS82R.
|
 |
Figure 7.
Fig. 7. Histogram of the hydrogenbond distance beteen acceptor and
donor within 2.4 and 3.4 A for 0T (gray bars) and cS82R (white
bars). (a) The proteinprotein interaction. (b) The proteinwater
interaction. (c) The waterwater interacton.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(1996,
52,
623-630)
copyright 1996.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Ă.‰.Gráczer,
A.Merli,
R.K.Singh,
M.Karuppasamy,
P.Závodszky,
M.S.Weiss,
and
M.Vas
(2011).
Atomic level description of the domain closure in a dimeric enzyme: thermus thermophilus 3-isopropylmalate dehydrogenase.
|
| |
Mol Biosyst,
7,
1646-1659.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Weik,
and
J.P.Colletier
(2010).
Temperature-dependent macromolecular X-ray crystallography.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
437-446.
|
 |
|
|
|
|
 |
J.Funahashi,
K.Takano,
Y.Yamagata,
and
K.Yutani
(2002).
Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
|
| |
J Biol Chem,
277,
21792-21800.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

