 |
PDBsum entry 1ws3

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ws3
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Urate oxidase from aspergillus flavus complexed with uracil
|
|
Structure:
|
 |
Uricase. Chain: a, b, c, d. Synonym: urate oxidase. Engineered: yes
|
|
Source:
|
 |
Aspergillus flavus. Organism_taxid: 5059. Expressed in: saccharomyces cerevisiae. Expression_system_taxid: 4932
|
|
Biol. unit:
|
 |
Tetramer (from
)
|
|
Resolution:
|
 |
|
3.20Å
|
R-factor:
|
0.146
|
R-free:
|
0.200
|
|
|
Authors:
|
 |
P.Retailleau,N.Colloc'H,D.Vivares,F.Bonnete,B.Castro,M.El Hajji, T.Prange
|
Key ref:

|
 |
P.Retailleau
et al.
(2005).
Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
Acta Crystallogr D Biol Crystallogr,
61,
218-229.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
29-Oct-04
|
Release date:
|
22-Mar-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q00511
(URIC_ASPFL) -
Uricase from Aspergillus flavus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
302 a.a.
296 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.7.3.3
- factor independent urate hydroxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
urate + O2 + H2O = 5-hydroxyisourate + H2O2
|
 |
 |
 |
 |
 |
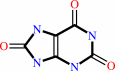 urate
urate
|
+
|
 O2
O2
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 66.67% similarity
|
=
|
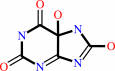 5-hydroxyisourate
5-hydroxyisourate
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Copper
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
61:218-229
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Urate oxidase from Aspergillus flavus: new crystal-packing contacts in relation to the content of the active site.
|
|
P.Retailleau,
N.Colloc'h,
D.Vivarès,
F.Bonneté,
B.Castro,
M.El Hajji,
T.Prangé.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Urate oxidase from Aspergillus flavus (uricase or Uox; EC 1.7.3.3) is a 135 kDa
homotetramer with a subunit consisting of 301 amino acids. It catalyses the
first step of the degradation of uric acid into allantoin. The structure of the
extracted enzyme complexed with a purine-type inhibitor (8-azaxanthin) had been
solved from high-resolution X-ray diffraction of I222 crystals. Expression of
the recombinant enzyme in Saccharomyces cerevisiae followed by a new
purification procedure allowed the crystallization of both unliganded and
liganded enzymes utilizing the same conditions but in various crystal forms.
Here, four different crystal forms of Uox are analyzed. The diversity of the Uox
crystal forms appears to depend strongly on the chemicals used as inhibitors. In
the presence of uracil and 5,6-diaminouracil crystals usually belong to the
trigonal space group P3(1)21, the asymmetric unit (AU) of which contains one
tetramer of Uox (four subunits). Chemical oxidation of 5,6-diaminouracil within
the protein may occur, leading to the canonical (I222) packing with one subunit
per AU. Coexistence of two crystal forms, P2(1) with two tetramers per AU and
I222, was found in the same crystallization drop containing another inhibitor,
guanine. Finally, a fourth form, P2(1)2(1)2 with one tetramer per AU, resulted
fortuitously in the presence of cymelarsan, an additive. Of all the reported
forms, the I222 crystal forms present by far the best X-ray diffraction
resolution (approximately 1.6 angstroms resolution compared with 2.3-3.2
angstroms for the other forms). The various structures and contacts in all
crystalline lattices are compared. The backbones are essentially conserved
except for the region near the active site. Its location at the dimer interface
is thus likely to be at the origin of the crystal contact changes as a response
to the various bound inhibitors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1
View of the active site: the omit map is contoured at the 1.5 [118][sigma] level around
the 5-amino-6-nitrouracil with the model superimposed. Graphics were created using
MOLSCRIPT (Kraulis, 1991[119] [Kraulis, P. E. (1991). J. Appl. Cryst. 24,
946-950.]-[120][bluearr.gif] ) and rendered using RASTER3D (Merritt & Murphy, 1994[121]
[Merritt, E. A. & Murphy, M. E. P. (1994). Acta Cryst. D50, 869-873.]-[122][bluearr.gif]
).
|
 |
Figure 2.
Figure 2
The reaction pathway from uric acid to 5-hydroxyisourate (middle) and the analogues used.
Note that uracil (and derivatives) standard numberings differ compared with uric acid
derivatives. *, Kahn & Tipton (1997[159] [Kahn, K. & Tipton, P. A. (1997). Biochemistry,
36, 4731-4738.]-[160][bluearr.gif] ); **, Pfleiderer (1974[161] [Pfleiderer, W. (1974).
Liebigs Ann. Chem. 12, 2030-2045.]-[162][bluearr.gif] ).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2005,
61,
218-229)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.Gabison,
C.Chopard,
N.Colloc'h,
F.Peyrot,
B.Castro,
M.E.Hajji,
M.Altarsha,
G.Monard,
M.Chiadmi,
and
T.Prangé
(2011).
X-ray, ESR, and quantum mechanics studies unravel a spin well in the cofactor-less urate oxidase.
|
| |
Proteins,
79,
1964-1976.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Pompidor,
O.Maury,
J.Vicat,
and
R.Kahn
(2010).
A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
762-769.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Collings,
Y.Watier,
M.Giffard,
S.Dagogo,
R.Kahn,
F.Bonneté,
J.P.Wright,
A.N.Fitch,
and
I.Margiolaki
(2010).
Polymorphism of microcrystalline urate oxidase from Aspergillus flavus.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
539-548.
|
 |
|
|
|
|
 |
L.Gabison,
M.Chiadmi,
M.El Hajji,
B.Castro,
N.Colloc'h,
and
T.Prangé
(2010).
Near-atomic resolution structures of urate oxidase complexed with its substrate and analogues: the protonation state of the ligand.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
714-724.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Oksanen,
M.P.Blakeley,
F.Bonneté,
M.T.Dauvergne,
F.Dauvergne,
and
M.Budayova-Spano
(2009).
Large crystal growth by thermal control allows combined X-ray and neutron crystallographic studies to elucidate the protonation states in Aspergillus flavus urate oxidase.
|
| |
J R Soc Interface,
6,
S599-S610.
|
 |
|
|
|
|
 |
L.Gabison,
T.Prangé,
N.Colloc'h,
M.El Hajji,
B.Castro,
and
M.Chiadmi
(2008).
Structural analysis of urate oxidase in complex with its natural substrate inhibited by cyanide: mechanistic implications.
|
| |
BMC Struct Biol,
8,
32.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.K.Jung,
Y.Lee,
S.G.Park,
B.C.Park,
G.H.Kim,
and
S.Rhee
(2006).
Structural and functional analysis of PucM, a hydrolase in the ureide pathway and a member of the transthyretin-related protein family.
|
| |
Proc Natl Acad Sci U S A,
103,
9790-9795.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

