 |
PDBsum entry 1w2f

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Human inositol (1,4,5)-trisphosphate 3-kinase substituted with selenomethionine
|
|
Structure:
|
 |
Inositol-trisphosphate 3-kinase a. Chain: a, b. Fragment: catalytic domain, residues 186-461. Synonym: inositol 1,4,5-trisphosphate 3-kinase, ip3-3k. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562. Expression_system_variant: de3.
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.204
|
R-free:
|
0.241
|
|
|
Authors:
|
 |
B.Gonzalez,M.J.Schell,R.F.Irvine,R.L.Williams
|
Key ref:

|
 |
B.González
et al.
(2004).
Structure of a human inositol 1,4,5-trisphosphate 3-kinase: substrate binding reveals why it is not a phosphoinositide 3-kinase.
Mol Cell,
15,
689-701.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
01-Jul-04
|
Release date:
|
09-Sep-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P23677
(IP3KA_HUMAN) -
Inositol-trisphosphate 3-kinase A from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
461 a.a.
276 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.127
- inositol-trisphosphate 3-kinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Phosphate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1D-myo-inositol 1,4,5-trisphosphate + ATP = 1D-myo-inositol 1,3,4,5- tetrakisphosphate + ADP + H+
|
 |
 |
 |
 |
 |
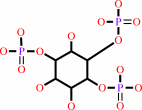 1D-myo-inositol 1,4,5-trisphosphate
1D-myo-inositol 1,4,5-trisphosphate
|
+
|
 ATP
ATP
|
=
|
 1D-myo-inositol 1,3,4,5- tetrakisphosphate
1D-myo-inositol 1,3,4,5- tetrakisphosphate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Ca(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Mol Cell
15:689-701
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of a human inositol 1,4,5-trisphosphate 3-kinase: substrate binding reveals why it is not a phosphoinositide 3-kinase.
|
|
B.González,
M.J.Schell,
A.J.Letcher,
D.B.Veprintsev,
R.F.Irvine,
R.L.Williams.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Mammalian cells produce a variety of inositol phosphates (InsPs), including
Ins(1,4,5)P3 that serves both as a second messenger and as a substrate for
inositol polyphosphate kinases (IPKs), which further phosphorylate it. We report
the structure of an IPK, the human Ins(1,4,5)P3 3-kinase-A, both free and in
complexes with substrates and products. This enzyme catalyzes transfer of a
phosphate from ATP to the 3-OH of Ins(1,4,5)P3, and its X-ray crystal structure
provides a template for understanding a broad family of InsP kinases. The
catalytic domain consists of three lobes. The N and C lobes bind ATP and
resemble protein and lipid kinases, despite insignificant sequence similarity.
The third lobe binds inositol phosphate and is a unique four-helix insertion in
the C lobe. This lobe embraces all of the phosphates of Ins(1,4,5)P3 in a
positively charged pocket, explaining the enzyme's substrate specificity and its
inability to phosphorylate PtdIns(4,5)P2, the membrane-resident analog of
Ins(1,4,5)P3.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. The Overall Fold of IP[3]-3K Catalytic Domain(A)
A ribbon diagram of IP[3]-3K in a complex with
Mn^2+/AMPPNP/Ins(1,4,5)P[3]. The N lobe is colored orange, the C
lobe yellow, the IP lobe purple, and the hinge green. The
conserved IDFG, GSSLL, and DxK motifs are colored pink, cyan,
and brown, respectively. This and all other molecular
illustrations were prepared with PYMOL.(B) A topology diagram of
the catalytic domain. Helix α0[N] is ordered only in the
absence of substrates or products. The dashed arrow indicates an
additional strand that would be characteristic of the N lobes of
PKs.(C) Ribbon diagrams of the catalytic domains of PI3Kγ and
PIPKIIβ with N and C lobes colored as in (A).
|
 |
Figure 3.
Figure 3. Substrate Binding by IP[3]-3K(A) A view of the
active site showing the ATP analog and Ins(1,4,5)P[3] bound to
the enzyme. The subdomains and motifs are colored as in Figure
1. Side chains interacting with the ligands are shown as sticks
and ligand-bound water molecules as white spheres.(B) A
schematic of the interactions with the ligands made with
LIGPLOT.(C) A surface illustration of the IP[3]-3K catalytic
domain colored by electrostatic potential with negatively
charged regions red and positive blue. The bound AMPPNP is shown
as green sticks and the Ins(1,4,5)P[3] as yellow sticks.(D) A
close-up of the ATP binding pocket in the presence of AMPPNP
(left) and in its absence (right), showing Trp188 and His194
from the putative auto-inhibitory region mimicking the
interactions formed by ATP.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Mol Cell
(2004,
15,
689-701)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
B.González,
J.I.Baños-Sanz,
M.Villate,
C.A.Brearley,
and
J.Sanz-Aparicio
(2010).
Inositol 1,3,4,5,6-pentakisphosphate 2-kinase is a distant IPK member with a singular inositide binding site for axial 2-OH recognition.
|
| |
Proc Natl Acad Sci U S A,
107,
9608-9613.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.E.Cassidy,
and
W.N.Setzer
(2010).
Cancer-relevant biochemical targets of cytotoxic Lonchocarpus flavonoids: a molecular docking analysis.
|
| |
J Mol Model,
16,
311-326.
|
 |
|
|
|
|
 |
K.Sauer,
and
M.P.Cooke
(2010).
Regulation of immune cell development through soluble inositol-1,3,4,5-tetrakisphosphate.
|
| |
Nat Rev Immunol,
10,
257-271.
|
 |
|
|
|
|
 |
M.J.Schell
(2010).
Inositol trisphosphate 3-kinases: focus on immune and neuronal signaling.
|
| |
Cell Mol Life Sci,
67,
1755-1778.
|
 |
|
|
|
|
 |
A.R.Kinjo,
and
H.Nakamura
(2009).
Comprehensive structural classification of ligand-binding motifs in proteins.
|
| |
Structure,
17,
234-246.
|
 |
|
|
|
|
 |
S.M.Onnebo,
and
A.Saiardi
(2009).
Inositol pyrophosphates modulate hydrogen peroxide signalling.
|
| |
Biochem J,
423,
109-118.
|
 |
|
|
|
|
 |
U.Padmanabhan,
D.E.Dollins,
P.C.Fridy,
J.D.York,
and
C.P.Downes
(2009).
Characterization of a Selective Inhibitor of Inositol Hexakisphosphate Kinases: USE IN DEFINING BIOLOGICAL ROLES AND METABOLIC RELATIONSHIPS OF INOSITOL PYROPHOSPHATES.
|
| |
J Biol Chem,
284,
10571-10582.
|
 |
|
|
|
|
 |
A.Chakraborty,
M.A.Koldobskiy,
K.M.Sixt,
K.R.Juluri,
A.K.Mustafa,
A.M.Snowman,
D.B.van Rossum,
R.L.Patterson,
and
S.H.Snyder
(2008).
HSP90 regulates cell survival via inositol hexakisphosphate kinase-2.
|
| |
Proc Natl Acad Sci U S A,
105,
1134-1139.
|
 |
|
|
|
|
 |
A.R.Alcázar-Román,
and
S.R.Wente
(2008).
Inositol polyphosphates: a new frontier for regulating gene expression.
|
| |
Chromosoma,
117,
1.
|
 |
|
|
|
|
 |
P.Draskovic,
A.Saiardi,
R.Bhandari,
A.Burton,
G.Ilc,
M.Kovacevic,
S.H.Snyder,
and
M.Podobnik
(2008).
Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups.
|
| |
Chem Biol,
15,
274-286.
|
 |
|
|
|
|
 |
R.Bhandari,
K.R.Juluri,
A.C.Resnick,
and
S.H.Snyder
(2008).
Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis.
|
| |
Proc Natl Acad Sci U S A,
105,
2349-2353.
|
 |
|
|
|
|
 |
A.Rosenhouse-Dantsker,
and
D.E.Logothetis
(2007).
Molecular characteristics of phosphoinositide binding.
|
| |
Pflugers Arch,
455,
45-53.
|
 |
|
|
|
|
 |
J.H.Choi,
J.Williams,
J.Cho,
J.R.Falck,
and
S.B.Shears
(2007).
Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress.
|
| |
J Biol Chem,
282,
30763-30775.
|
 |
|
|
|
|
 |
S.M.Lloyd-Burton,
J.C.Yu,
R.F.Irvine,
and
M.J.Schell
(2007).
Regulation of inositol 1,4,5-trisphosphate 3-kinases by calcium and localization in cells.
|
| |
J Biol Chem,
282,
9526-9535.
|
 |
|
|
|
|
 |
M.M.Nalaskowski,
S.Windhorst,
M.C.Stockebrand,
and
G.W.Mayr
(2006).
Subcellular localisation of human inositol 1,4,5-trisphosphate 3-kinase C: species-specific use of alternative export sites for nucleo-cytoplasmic shuttling indicates divergent roles of the catalytic and N-terminal domains.
|
| |
Biol Chem,
387,
583-593.
|
 |
|
|
|
|
 |
R.F.Irvine,
S.M.Lloyd-Burton,
J.C.Yu,
A.J.Letcher,
and
M.J.Schell
(2006).
The regulation and function of inositol 1,4,5-trisphosphate 3-kinases.
|
| |
Adv Enzyme Regul,
46,
314-323.
|
 |
|
|
|
|
 |
S.Morgan-Lappe,
K.W.Woods,
Q.Li,
M.G.Anderson,
M.E.Schurdak,
Y.Luo,
V.L.Giranda,
S.W.Fesik,
and
J.D.Leverson
(2006).
RNAi-based screening of the human kinome identifies Akt-cooperating kinases: a new approach to designing efficacious multitargeted kinase inhibitors.
|
| |
Oncogene,
25,
1340-1348.
|
 |
|
|
|
|
 |
W.Holmes,
and
G.Jogl
(2006).
Crystal structure of inositol phosphate multikinase 2 and implications for substrate specificity.
|
| |
J Biol Chem,
281,
38109-38116.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.C.Resnick,
A.M.Snowman,
B.N.Kang,
K.J.Hurt,
S.H.Snyder,
and
A.Saiardi
(2005).
Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity.
|
| |
Proc Natl Acad Sci U S A,
102,
12783-12788.
|
 |
|
|
|
|
 |
A.Poinas,
K.Backers,
A.M.Riley,
S.J.Mills,
C.Moreau,
B.V.Potter,
and
C.Erneux
(2005).
Interaction of the catalytic domain of inositol 1,4,5-trisphosphate 3-kinase A with inositol phosphate analogues.
|
| |
Chembiochem,
6,
1449-1457.
|
 |
|
|
|
|
 |
G.W.Mayr,
S.Windhorst,
and
K.Hillemeier
(2005).
Antiproliferative plant and synthetic polyphenolics are specific inhibitors of vertebrate inositol-1,4,5-trisphosphate 3-kinases and inositol polyphosphate multikinase.
|
| |
J Biol Chem,
280,
13229-13240.
|
 |
|
|
|
|
 |
H.J.Xia,
and
G.Yang
(2005).
Inositol 1,4,5-trisphosphate 3-kinases: functions and regulations.
|
| |
Cell Res,
15,
83-91.
|
 |
|
|
|
|
 |
R.F.Irvine
(2005).
Inositide evolution - towards turtle domination?
|
| |
J Physiol,
566,
295-300.
|
 |
|
|
|
|
 |
S.Cheek,
K.Ginalski,
H.Zhang,
and
N.V.Grishin
(2005).
A comprehensive update of the sequence and structure classification of kinases.
|
| |
BMC Struct Biol,
5,
6.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

