 |
PDBsum entry 1vz5

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1vz5
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.11.77
- alkyl sulfatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a primary linear alkyl sulfate ester + 2-oxoglutarate + O2 = an aldehyde + sulfate + succinate + CO2 + H+
|
 |
 |
 |
 |
 |
primary linear alkyl sulfate ester
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
 O2
O2
|
=
|
 aldehyde
aldehyde
|
+
|
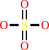 sulfate
sulfate
|
+
|
 succinate
succinate
|
+
|
CO2
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
280:5716-5723
(2005)
|
|
PubMed id:
|
|
|
|

|
| |
|
Succinate complex crystal structures of the alpha-ketoglutarate-dependent dioxygenase AtsK: steric aspects of enzyme self-hydroxylation.
|
|
I.Müller,
C.Stückl,
J.Wakeley,
M.Kertesz,
I.Usón.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The alkylsulfatase AtsK from Pseudomonas putida S-313 is a member of the
non-heme iron(II)-alpha-ketoglutarate-dependent dioxygenase superfamily. In the
initial step of their catalytic cycle, enzymes belonging to this widespread and
versatile family coordinate molecular oxygen to the iron center in the active
site. The subsequent decarboxylation of the cosubstrate alpha-ketoglutarate
yields carbon dioxide, succinate, and a highly reactive ferryl (IV) species,
which is required for substrate oxidation via a complex mechanism involving the
transfer of radical species. Non-productive activation of oxygen may lead to
harmful side reactions; therefore, such enzymes need an effective built-in
protection mechanism. One of the ways of controlling undesired side reactions is
the self-hydroxylation of an aromatic side chain, which leads to an irreversibly
inactivated species. Here we describe the crystal structure of the
alkylsulfatase AtsK in complexes with succinate and with Fe(II)/succinate. In
the crystal structure of the AtsK-Fe(II)-succinate complex, the side chain of
Tyr(168) is co-ordinated to the iron, suggesting that Tyr(168) is the target of
enzyme self-hydroxylation. This is the first structural study of an
Fe(II)-alpha-ketoglutarate-dependent dioxygenase that presents an aromatic side
chain coordinated to the metal center, thus allowing structural insight into
this protective mechanism of enzyme self-inactivation.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
FIG. 1. Alternative pathways for the reaction of the
protein complex with oxygen in presence and absence of
substrate. In the first step of the catalytic mechanism of all
non-heme iron(II)-  KG dependent
dioxygenases, iron and the cosubstrate KG dependent
dioxygenases, iron and the cosubstrate  KG coordinate to the
protein. In the next step, the substrate molecule approaches the
active site (on the left), thereby displacing a water molecule
from the metal center and liberating a coordinatively
unsaturated iron atom. This facilitates dioxygen binding in the
next step. One oxygen atom of O[2] is transferred to the
cosubstrate, yielding succinate and carbon dioxide as reaction
products. The iron is thereby oxidized, and a ferryl Fe(IV)=O
species is formed, which then hydroxylates the substrate via a
radical intermediate. In the absence of substrate, coordination
of a dioxygen molecule to the iron(II)· KG coordinate to the
protein. In the next step, the substrate molecule approaches the
active site (on the left), thereby displacing a water molecule
from the metal center and liberating a coordinatively
unsaturated iron atom. This facilitates dioxygen binding in the
next step. One oxygen atom of O[2] is transferred to the
cosubstrate, yielding succinate and carbon dioxide as reaction
products. The iron is thereby oxidized, and a ferryl Fe(IV)=O
species is formed, which then hydroxylates the substrate via a
radical intermediate. In the absence of substrate, coordination
of a dioxygen molecule to the iron(II)·  KG
complex can take place (on the right). In a self-protecting
mechanism, one possible reaction pathway of the ferryl species
formed after the decarboxylation of the KG
complex can take place (on the right). In a self-protecting
mechanism, one possible reaction pathway of the ferryl species
formed after the decarboxylation of the  KG is the reaction with
an amino acid side chain such as tryptophan or tyrosine, as
shown for the KG is the reaction with
an amino acid side chain such as tryptophan or tyrosine, as
shown for the  KG-dependent
dioxygenases TfdA, AlkB, or TauD. As an alternative to this
self-hydroxylation mechanism, the ferryl intermediate could
react with a second cosubstrate molecule. KG-dependent
dioxygenases TfdA, AlkB, or TauD. As an alternative to this
self-hydroxylation mechanism, the ferryl intermediate could
react with a second cosubstrate molecule.
|
 |
Figure 5.
FIG. 5. Active site region of the H[2]O-succinate-AtsK
complex (a) the Fe-succinate-AtsK complex (b). The amino acids
Ala^80 and His81 and Tyr166-Ala^169 of formerly disordered loops
participate in a hydrogen bond network in the
iron-succinate-AtsK complex that differs from that observed in
the succinate-AtsK complex. The new hydrogen bonds and the
relocation of  -sheet -sheet  4 result
in the formation of a lid over the active site of the
Fe-succinate-AtsK complex. 4 result
in the formation of a lid over the active site of the
Fe-succinate-AtsK complex.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2005,
280,
5716-5723)
copyright 2005.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Spiteller
(2010).
Is lipid peroxidation of polyunsaturated acids the only source of free radicals that induce aging and age-related diseases?
|
| |
Rejuvenation Res,
13,
91.
|
 |
|
|
|
|
 |
D.A.Small,
W.Chang,
F.Toghrol,
and
W.E.Bentley
(2007).
Comparative global transcription analysis of sodium hypochlorite, peracetic acid, and hydrogen peroxide on Pseudomonas aeruginosa.
|
| |
Appl Microbiol Biotechnol,
76,
1093-1105.
|
 |
|
|
|
|
 |
D.A.Small,
W.Chang,
F.Toghrol,
and
W.E.Bentley
(2007).
Toxicogenomic analysis of sodium hypochlorite antimicrobial mechanisms in Pseudomonas aeruginosa.
|
| |
Appl Microbiol Biotechnol,
74,
176-185.
|
 |
|
|
|
|
 |
V.Purpero,
and
G.R.Moran
(2007).
The diverse and pervasive chemistries of the alpha-keto acid dependent enzymes.
|
| |
J Biol Inorg Chem,
12,
587-601.
|
 |
|
|
|
|
 |
K.D.Koehntop,
S.Marimanikkuppam,
M.J.Ryle,
R.P.Hausinger,
and
L.Que
(2006).
Self-hydroxylation of taurine/alpha-ketoglutarate dioxygenase: evidence for more than one oxygen activation mechanism.
|
| |
J Biol Inorg Chem,
11,
63-72.
|
 |
|
|
|
|
 |
T.A.Müller,
M.I.Zavodszky,
M.Feig,
L.A.Kuhn,
and
R.P.Hausinger
(2006).
Structural basis for the enantiospecificities of R- and S-specific phenoxypropionate/alpha-ketoglutarate dioxygenases.
|
| |
Protein Sci,
15,
1356-1368.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

