 |
PDBsum entry 1vpd

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1vpd
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.60
- 2-hydroxy-3-oxopropionate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
(R)-glycerate + NADP+ = 2-hydroxy-3-oxopropanoate + NADPH + H+
|
|
2.
|
(R)-glycerate + NAD+ = 2-hydroxy-3-oxopropanoate + NADH + H+
|
|
 |
 |
 |
 |
 |
(R)-glycerate
Bound ligand (Het Group name = )
matches with 70.00% similarity
|
+
|
 NADP(+)
NADP(+)
|
=
|
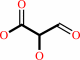 2-hydroxy-3-oxopropanoate
2-hydroxy-3-oxopropanoate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
(R)-glycerate
Bound ligand (Het Group name = )
matches with 70.00% similarity
|
+
|
 NAD(+)
NAD(+)
|
=
|
 2-hydroxy-3-oxopropanoate
2-hydroxy-3-oxopropanoate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Struct Funct Genomics
10:249-253
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
|
|
J.Osipiuk,
M.Zhou,
S.Moy,
F.Collart,
A.Joachimiak.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Tartronate semialdehyde reductases (TSRs), also known as
2-hydroxy-3-oxopropionate reductases, catalyze the reduction of tartronate
semialdehyde using NAD as cofactor in the final stage of D-glycerate
biosynthesis. These enzymes belong to family of structurally and mechanically
related beta-hydroxyacid dehydrogenases which differ in substrate specificity
and catalyze reactions in specific metabolic pathways. Here, we present the
crystal structure of GarR a TSR from Salmonella typhimurium determined by the
single-wavelength anomalous diffraction method and refined to 1.65 A resolution.
The active site of the enzyme contains L-tartrate which most likely mimics a
position of a glycerate which is a product of the enzyme reaction. The analysis
of the TSR structure shows also a putative NADPH binding site in the enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Mondal,
C.Nagao,
and
K.Mizuguchi
(2010).
Detecting subtle functional differences in ketopantoate reductase and related enzymes using a rule-based approach with sequence-structure homology recognition scores.
|
| |
Protein Eng Des Sel,
23,
859-869.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

