 |
PDBsum entry 1vje

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.4.1.21
- S-ribosylhomocysteine lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Autoinducer AI-2 Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-(5-deoxy-D-ribos-5-yl)-L-homocysteine = (S)-4,5-dihydroxypentane-2,3- dione + L-homocysteine
|
 |
 |
 |
 |
 |
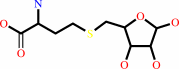 S-(5-deoxy-D-ribos-5-yl)-L-homocysteine
S-(5-deoxy-D-ribos-5-yl)-L-homocysteine
|
=
|
(S)-4,5-dihydroxypentane-2,3- dione
|
+
|
L-homocysteine
Bound ligand (Het Group name = )
matches with 70.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
9:527-537
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
|
|
H.A.Lewis,
E.B.Furlong,
B.Laubert,
G.A.Eroshkina,
Y.Batiyenko,
J.M.Adams,
M.G.Bergseid,
C.D.Marsh,
T.S.Peat,
W.E.Sanderson,
J.M.Sauder,
S.G.Buchanan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Quorum sensing is the mechanism by which bacteria control gene
expression in response to cell density. Two major quorum-sensing systems have
been identified, system 1 and system 2, each with a characteristic signaling
molecule (autoinducer-1, or AI-1, in the case of system 1, and AI-2 in system
2). The luxS gene is required for the AI-2 system of quorum sensing. LuxS and
AI-2 have been described in both Gram-negative and Gram-positive bacterial
species and have been shown to be involved in the expression of virulence genes
in several pathogens. RESULTS: The structure of the LuxS protein from three
different bacterial species with resolutions ranging from 1.8 A to 2.4 A has
been solved using an X-ray crystallographic structural genomics approach. The
structure of LuxS reported here is seen to have a new alpha-beta fold. In all
structures, an equivalent homodimer is observed. A metal ion identified as zinc
was seen bound to a Cys-His-His triad. Methionine was found bound to the protein
near the metal and at the dimer interface. CONCLUSIONS: These structures provide
support for a hypothesis that explains the in vivo action of LuxS. Specifically,
acting as a homodimer, the protein binds a methionine analog,
S-ribosylhomocysteine (SRH). The zinc atom is in position to cleave the ribose
ring in a step along the synthesis pathway of AI-2.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 6.
Figure 6. Ball and Stick and Ribbon Diagram of the
Substrate and Metal Binding SitesPotential hydrogen-bonding
partners for the methionine ligand are shown by white dotted
lines. Potential hydrogen bonding interactions with the zinc
atom are also shown

|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2001,
9,
527-537)
copyright 2001.
|
|
| |
Figure was
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Bhattacharyya,
and
S.Vishveshwara
(2010).
Elucidation of the conformational free energy landscape in H.pylori LuxS and its implications to catalysis.
|
| |
BMC Struct Biol,
10,
27.
|
 |
|
|
|
|
 |
B.Gopishetty,
J.Zhu,
R.Rajan,
A.J.Sobczak,
S.F.Wnuk,
C.E.Bell,
and
D.Pei
(2009).
Probing the catalytic mechanism of S-ribosylhomocysteinase (LuxS) with catalytic intermediates and substrate analogues.
|
| |
J Am Chem Soc,
131,
1243-1250.
|
 |
|
|
|
|
 |
M.Bhattacharyya,
and
S.Vishveshwara
(2009).
Functional correlation of bacterial LuxS with their quaternary associations: interface analysis of the structure networks.
|
| |
BMC Struct Biol,
9,
8.
|
 |
|
|
|
|
 |
M.Zhang,
X.D.Jiao,
Y.H.Hu,
and
L.Sun
(2009).
Attenuation of Edwardsiella tarda virulence by small peptides that interfere with LuxS/autoinducer type 2 quorum sensing.
|
| |
Appl Environ Microbiol,
75,
3882-3890.
|
 |
|
|
|
|
 |
M.Zhang,
K.Sun,
and
L.Sun
(2008).
Regulation of autoinducer 2 production and luxS expression in a pathogenic Edwardsiella tarda strain.
|
| |
Microbiology,
154,
2060-2069.
|
 |
|
|
|
|
 |
Y.Turovskiy,
D.Kashtanov,
B.Paskhover,
and
M.L.Chikindas
(2007).
Quorum sensing: fact, fiction, and everything in between.
|
| |
Adv Appl Microbiol,
62,
191-234.
|
 |
|
|
|
|
 |
C.Okada,
Y.Maegawa,
M.Yao,
and
I.Tanaka
(2006).
Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold.
|
| |
Proteins,
63,
1119-1122.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.C.Petersen,
N.A.Ahmed,
A.Naemi,
and
A.A.Scheie
(2006).
LuxS-mediated signalling in Streptococcus anginosus and its role in biofilm formation.
|
| |
Antonie Van Leeuwenhoek,
90,
109-121.
|
 |
|
|
|
|
 |
J.E.González,
and
N.D.Keshavan
(2006).
Messing with bacterial quorum sensing.
|
| |
Microbiol Mol Biol Rev,
70,
859-875.
|
 |
|
|
|
|
 |
S.C.De Keersmaecker,
K.Sonck,
and
J.Vanderleyden
(2006).
Let LuxS speak up in AI-2 signaling.
|
| |
Trends Microbiol,
14,
114-119.
|
 |
|
|
|
|
 |
A.Vendeville,
K.Winzer,
K.Heurlier,
C.M.Tang,
and
K.R.Hardie
(2005).
Making 'sense' of metabolism: autoinducer-2, LuxS and pathogenic bacteria.
|
| |
Nat Rev Microbiol,
3,
383-396.
|
 |
|
|
|
|
 |
J.Badger,
J.M.Sauder,
J.M.Adams,
S.Antonysamy,
K.Bain,
M.G.Bergseid,
S.G.Buchanan,
M.D.Buchanan,
Y.Batiyenko,
J.A.Christopher,
S.Emtage,
A.Eroshkina,
I.Feil,
E.B.Furlong,
K.S.Gajiwala,
X.Gao,
D.He,
J.Hendle,
A.Huber,
K.Hoda,
P.Kearins,
C.Kissinger,
B.Laubert,
H.A.Lewis,
J.Lin,
K.Loomis,
D.Lorimer,
G.Louie,
M.Maletic,
C.D.Marsh,
I.Miller,
J.Molinari,
H.J.Muller-Dieckmann,
J.M.Newman,
B.W.Noland,
B.Pagarigan,
F.Park,
T.S.Peat,
K.W.Post,
S.Radojicic,
A.Ramos,
R.Romero,
M.E.Rutter,
W.E.Sanderson,
K.D.Schwinn,
J.Tresser,
J.Winhoven,
T.A.Wright,
L.Wu,
J.Xu,
and
T.J.Harris
(2005).
Structural analysis of a set of proteins resulting from a bacterial genomics project.
|
| |
Proteins,
60,
787-796.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.M.Pappas,
C.L.Weingart,
and
S.C.Winans
(2004).
Chemical communication in proteobacteria: biochemical and structural studies of signal synthases and receptors required for intercellular signalling.
|
| |
Mol Microbiol,
53,
755-769.
|
 |
|
|
|
|
 |
F.Schafer,
A.Schafer,
and
K.Steinert
(2002).
A highly specific system for efficient enzymatic removal of tags from recombinant proteins.
|
| |
J Biomol Tech,
13,
158-171.
|
 |
|
|
|
|
 |
F.Schafer,
U.Romer,
M.Emmerlich,
J.Blumer,
H.Lubenow,
and
K.Steinert
(2002).
Automated high-throughput purification of 6xHis-tagged proteins.
|
| |
J Biomol Tech,
13,
131-142.
|
 |
|
|
|
|
 |
L.Stewart,
R.Clark,
and
C.Behnke
(2002).
High-throughput crystallization and structure determination in drug discovery.
|
| |
Drug Discov Today,
7,
187-196.
|
 |
|
|
|
|
 |
M.P.DeLisa,
and
W.E.Bentley
(2002).
Bacterial autoinduction: looking outside the cell for new metabolic engineering targets.
|
| |
Microb Cell Fact,
1,
5.
|
 |
|
|
|
|
 |
O.M.Cloak,
B.T.Solow,
C.E.Briggs,
C.Y.Chen,
and
P.M.Fratamico
(2002).
Quorum sensing and production of autoinducer-2 in Campylobacter spp., Escherichia coli O157:H7, and Salmonella enterica serovar Typhimurium in foods.
|
| |
Appl Environ Microbiol,
68,
4666-4671.
|
 |
|
|
|
|
 |
W.T.Watson,
T.D.Minogue,
D.L.Val,
S.B.von Bodman,
and
M.E.Churchill
(2002).
Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing.
|
| |
Mol Cell,
9,
685-694.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.N.Lawrence
(2001).
News in brief.
|
| |
Drug Discov Today,
6,
759-762.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

