 |
PDBsum entry 1vft

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.1.1
- alanine racemase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-alanine = D-alanine
|
 |
 |
 |
 |
 |
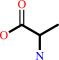 L-alanine
L-alanine
|
=
|
 D-alanine
D-alanine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
DCS)
matches with 65.22% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
279:46153-46161
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
|
|
M.Noda,
Y.Matoba,
T.Kumagai,
M.Sugiyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Alanine racemase (ALR), an enzyme that catalyzes the interconversion of Ala
enantiomers, is essential for the synthesis of the bacterial cell wall. We have
shown that it is harder to inhibit the catalytic activity of ALR from
D-cycloserine (DCS)-producing Streptomyces lavendulae than that from Escherichia
coli by DCS. To obtain structural evidence for the fact that Streptomyces ALR
displays resistance to DCS, we determined the precise nature of the x-ray
crystal structures of the cycloserine-free and cycloserine enantiomer-bound
forms of Streptomyces ALR at high resolutions. Streptomyces ALR takes a dimer
structure, which is formed by interactions between the N-terminal domain of one
monomer with the C-terminal domain of its partner. Each of the two active sites
of ALR, which is generated as a result of the formation of the dimer structure,
is composed of pyridoxal 5'-phosphate (PLP), the PLP-binding residue Lys(38),
and the amino acids in the immediate environment of the pyridoxal cofactor. The
current model suggests that each active site of Streptomyces ALR maintains a
larger space and takes a more rigid conformation than that of Bacillus
stearothermophilus ALR determined previously. Furthermore, we show that
Streptomyces ALR results in a slow conversion to a final form of a pyridoxal
derivative arising from either isomer of cycloserine, which inhibits the
catalytic activity noncompetitively. In fact, the slow conversion is confirmed
by the fact that each enzyme bound cycloserine derivative, which is bound to
PLP, takes an asymmetric structure.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 6.
FIG. 6. Active site around the DCS- or LCS-bound PLP
derivative. DCS-bound derivatives at sites A and B are shown in
a and b, respectively. The LCS-bound derivatives at sites A and
B are shown in c and d, respectively. The carbon atoms of the
residues from each monomer are shown in orange and cyan. The
carbon atoms of the residues from the PLP derivative, designated
SCP, are shown in purple.
|
 |
Figure 7.
FIG. 7. Proposed mechanism of inactivation for ALR by DCS
or LCS. DCS and LCS are converted to an identical final aromatic
adduct through transamination, transamination, and
tautomerization steps. The DCS- or LSC-bound PLP intermediate
following the transamination step can be racemized to each
counterpart by the same reaction as used for the true substrates.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2004,
279,
46153-46161)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.R.Scaletti,
S.R.Luckner,
and
K.L.Krause
(2012).
Structural features and kinetic characterization of alanine racemase from Staphylococcus aureus (Mu50).
|
| |
Acta Crystallogr D Biol Crystallogr,
68,
82-92.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Ju,
S.Xu,
Y.Furukawa,
Y.Zhang,
H.Misono,
T.Minamino,
K.Namba,
B.Zhao,
and
K.Ohnishi
(2011).
Correlation between catalytic activity and monomer-dimer equilibrium of bacterial alanine racemases.
|
| |
J Biochem,
149,
83-89.
|
 |
|
|
|
|
 |
J.Lowther,
B.A.Yard,
K.A.Johnson,
L.G.Carter,
V.T.Bhat,
M.C.Raman,
D.J.Clarke,
B.Ramakers,
S.A.McMahon,
J.H.Naismith,
and
D.J.Campopiano
(2010).
Inhibition of the PLP-dependent enzyme serine palmitoyltransferase by cycloserine: evidence for a novel decarboxylative mechanism of inactivation.
|
| |
Mol Biosyst,
6,
1682-1693.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.M.Couñago,
M.Davlieva,
U.Strych,
R.E.Hill,
and
K.L.Krause
(2009).
Biochemical and structural characterization of alanine racemase from Bacillus anthracis (Ames).
|
| |
BMC Struct Biol,
9,
53.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Wu,
T.Hu,
L.Zhang,
J.Chen,
J.Du,
J.Ding,
H.Jiang,
and
X.Shen
(2008).
Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis.
|
| |
Protein Sci,
17,
1066-1076.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Liu,
P.D.Fortin,
and
C.T.Walsh
(2008).
Andrimid producers encode an acetyl-CoA carboxyltransferase subunit resistant to the action of the antibiotic.
|
| |
Proc Natl Acad Sci U S A,
105,
13321-13326.
|
 |
|
|
|
|
 |
J.Ju,
K.Yokoigawa,
H.Misono,
and
K.Ohnishi
(2005).
Cloning of alanine racemase genes from Pseudomonas fluorescens strains and oligomerization states of gene products expressed in Escherichia coli.
|
| |
J Biosci Bioeng,
100,
409-417.
|
 |
|
|
|
|
 |
L.Lehtiö,
I.Fabrichniy,
T.Hansen,
P.Schönheit,
and
A.Goldman
(2005).
Unusual twinning in an acetyl coenzyme A synthetase (ADP-forming) from Pyrococcus furiosus.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
350-354.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

