 |
PDBsum entry 1v7r

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.6.1.66
- XTP/dITP diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
XTP + H2O = XMP + diphosphate + H+
|
|
2.
|
dITP + H2O = dIMP + diphosphate + H+
|
|
3.
|
ITP + H2O = IMP + diphosphate + H+
|
|
 |
 |
 |
 |
 |
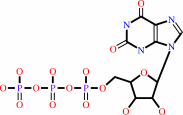 XTP
XTP
|
+
|
H2O
|
=
|
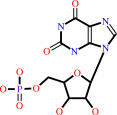 XMP
XMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
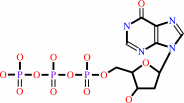 dITP
dITP
|
+
|
H2O
|
=
|
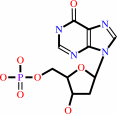 dIMP
dIMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
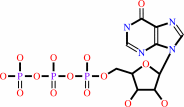 ITP
ITP
|
+
|
H2O
|
=
|
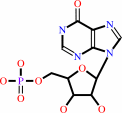 IMP
IMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
375:1013-1025
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of dimeric nonstandard nucleotide triphosphate pyrophosphatase from Pyrococcus horikoshii OT3: functional significance of interprotomer conformational changes.
|
|
N.K.Lokanath,
K.J.Pampa,
K.Takio,
N.Kunishima.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Nonstandard nucleotide triphosphate pyrophosphatase (NTPase) can efficiently
hydrolyze nonstandard purine nucleotides in the presence of divalent cations.
The crystal structures of the NTPase from Pyrococcus horikoshii OT3 (PhNTPase)
have been determined in two unliganded forms and in three liganded forms with
inosine 5'-monophosphate (IMP), ITP and Mn(2+), which visualize the recognition
of these ligands unambiguously. The overall structure of PhNTPase is similar to
that of previously reported crystal structures of the NTPase from Methanococcus
jannaschii and the human ITPase. They share a similar protomer folding with two
domains and a similar homodimeric quaternary structure. The dimeric interface of
NTPase is well conserved, and the dimeric state might be important to constitute
the active site of this enzyme. A conformational analysis of the five snapshots
of PhNTPase structures using the multiple superposition method reveals that IMP,
ITP and Mn(2+) bind to the active site without inducing large local
conformational changes, indicating that a combination of interdomain and
interprotomer rigid-body shifts mainly describes the conformational change of
PhNTPase. The interdomain and interprotomer conformations of the ITP liganded
form are essentially the same as those observed in the unliganded form 1,
indicating that ITP binding to PhNTPase in solution may follow the selection
mode in which ITP binds to the subunit that happens to be in the conformation
observed in the unliganded form 1. In contrast to the human ITPase inducing a
large domain closure upon ITP binding, the interdomain active site cleft is
generally closed in PhNTPase and only the IMP binding form shows a remarkable
domain opening by 14 degrees only in the B subunit. The interprotomer rigid-body
rotation of PhNTPase has a tendency to keep the dimeric 2-fold symmetry, which
is also true in human ITPase, thereby suggesting its relevance to the positive
cooperativity of the dimeric NTPases. The exception of this rule is observed in
the IMP liganded form in which the dimeric 2-fold symmetry is broken by a 3
degrees interprotomer rotation in an unusual direction. A combination of the
exceptional interdomain and interprotomer relocations is most likely the reason
for the observed asymmetric IMP binding that might be necessary for PhNTPase to
release the reaction product IMP.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. Structural superposition of the PhNTPase protomer
(magenta) with the MjNTPase (yellow) and human ITPase (green).
|
 |
Figure 5.
Fig. 5. Recognition of ligands in PhNTPase. Flat
representation of (a) IMP and (b) ITP binding sites. (c) Metal
binding site. The polar interactions are denoted by dashed lines
with distances in angstrom. Conserved residues and water
molecules are labeled. (d) Molecular surface representation
showing the IMP binding pocket.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2008,
375,
1013-1025)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Rlepokura,
and
R.Petrus
(2010).
Nucleotide-amino acid interactions in the L-His-IMP.MeOH.H(2)O complex.
|
| |
Acta Crystallogr C,
66,
o265-o269.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

