 |
PDBsum entry 1v7c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.3.1
- threonine synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Threonine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
O-phospho-L-homoserine + H2O = L-threonine + phosphate
|
 |
 |
 |
 |
 |
 O-phospho-L-homoserine
O-phospho-L-homoserine
|
+
|
H2O
|
=
|
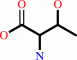 L-threonine
L-threonine
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
HEY)
matches with 53.57% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
278:46035-46045
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of threonine synthase from Thermus thermophilus HB8: conformational change, substrate recognition, and mechanism.
|
|
R.Omi,
M.Goto,
I.Miyahara,
H.Mizuguchi,
H.Hayashi,
H.Kagamiyama,
K.Hirotsu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Threonine synthase, which is a PLP-dependent enzyme, catalyzes the
beta,gamma-replacement reaction of l-homoserine phosphate to yield threonine and
inorganic phosphate. The three-dimensional structures of the enzyme from Thermus
thermophilus HB8 in its unliganded form and complexed with the substrate
analogue 2-amino-5-phosphonopentanoic acid have been determined at 2.15 and 2.0
A resolution, respectively. The complexed form, assigned as an enamine,
uncovered the interactions of the cofactor-analogue conjugate with the active
site residues. The binding of the substrate analogue induces a large
conformational change at the domain level. The small domain rotates by about 25
degrees and approaches the large domain to close the active site. The
complicated catalytic process of the enzyme has been elucidated based on the
complex structure to reveal the stereochemistry of the reaction and to present
the released inorganic phosphate as a possible catalyst to carry a proton to the
Cgamma atom of the substrate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 6.
FIG. 6. Schematic diagram showing hydrogen bond and salt
bridge interactions of the active site residues. The unliganded
tThrS (A) and tThrS·AP5 complex (B). Putative
interactions are shown by dotted lines if the acceptor and donor
are less than 3.3 Å apart.
|
 |
Figure 9.
FIG. 9. The proposed reaction mechanism of ThrS with the
substrate, HSerP. A, Michaelis complex; B, external aldimine; C,
carbanion intermediate; D, ketimine; E, enamine; F,  , ,  -unsaturated ketimine;
G, -unsaturated ketimine;
G,  , ,  -unsaturated aldimine;
H, external aldimine between PLP and threonine. -unsaturated aldimine;
H, external aldimine between PLP and threonine.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2003,
278,
46035-46045)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.E.Graham,
S.M.Taylor,
R.Z.Wolf,
and
S.C.Namboori
(2009).
Convergent evolution of coenzyme M biosynthesis in the Methanosarcinales: cysteate synthase evolved from an ancestral threonine synthase.
|
| |
Biochem J,
424,
467-478.
|
 |
|
|
|
|
 |
M.Goto,
T.Yamauchi,
N.Kamiya,
I.Miyahara,
T.Yoshimura,
H.Mihara,
T.Kurihara,
K.Hirotsu,
and
N.Esaki
(2009).
Crystal structure of a homolog of mammalian serine racemase from Schizosaccharomyces pombe.
|
| |
J Biol Chem,
284,
25944-25952.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Yamauchi,
M.Goto,
H.Y.Wu,
T.Uo,
T.Yoshimura,
H.Mihara,
T.Kurihara,
I.Miyahara,
K.Hirotsu,
and
N.Esaki
(2009).
Serine racemase with catalytically active lysinoalanyl residue.
|
| |
J Biochem,
145,
421-424.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.A.Azevedo,
M.Lancien,
and
P.J.Lea
(2006).
The aspartic acid metabolic pathway, an exciting and essential pathway in plants.
|
| |
Amino Acids,
30,
143-162.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

