 |
PDBsum entry 1v53

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1v53
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
The crystal structure of 3-isopropylmalate dehydrogenase from bacillus coagulans
|
|
Structure:
|
 |
3-isopropylmalate dehydrogenase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Bacillus coagulans. Organism_taxid: 1398. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.85Å
|
R-factor:
|
0.206
|
R-free:
|
0.250
|
|
|
Authors:
|
 |
K.Fujita,H.Minami,K.Suzuki,M.Tsunoda,T.Sekiguchi,R.Mizui,S.Tsuzaki, S.Nakamura,A.Takenaka
|
|
Key ref:
|
 |
K.Fujita
et al.
The crystal structure of 3-Isopropylmalate dehydrogenase from bacillus coagulans.
To be published,
.
|
 |
|
Date:
|
 |
|
20-Nov-03
|
Release date:
|
15-Feb-05
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P12010
(LEU3_WEICA) -
3-isopropylmalate dehydrogenase from Weizmannia coagulans
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
366 a.a.
356 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.85
- 3-isopropylmalate dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Leucine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3S)-3-isopropylmalate + NAD+ = 4-methyl-2-oxopentanoate + CO2 + NADH
|
 |
 |
 |
 |
 |
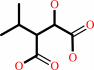 (2R,3S)-3-isopropylmalate
(2R,3S)-3-isopropylmalate
|
+
|
 NAD(+)
NAD(+)
|
=
|
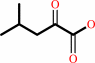 4-methyl-2-oxopentanoate
4-methyl-2-oxopentanoate
|
+
|
 CO2
CO2
|
+
|
 NADH
NADH
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

