 |
PDBsum entry 1v4p

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.7
- alanine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Ala) + L-alanine + ATP = L-alanyl-tRNA(Ala) + AMP + diphosphate
|
 |
 |
 |
 |
 |
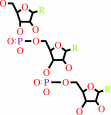 tRNA(Ala)
tRNA(Ala)
|
+
|
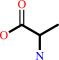 L-alanine
L-alanine
|
+
|
 ATP
ATP
|
=
|
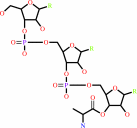 L-alanyl-tRNA(Ala)
L-alanyl-tRNA(Ala)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
62:1133-1137
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of alanyl-tRNA synthetase editing-domain homolog (PH0574) from a hyperthermophile, Pyrococcus horikoshii OT3 at 1.45 A resolution.
|
|
J.Ishijima,
Y.Uchida,
C.Kuroishi,
C.Tuzuki,
N.Takahashi,
N.Okazaki,
K.Yutani,
M.Miyano.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Multiple alignment of the editing domain of AlaRS and
ThrRS enzymes. Residues involved in coordination of the zinc ion
(red) and highly conserved residues (yellow) are indicated. The
secondary structure elements in the crystal structure of PH0574
(above) and ecThrRS (below) are shown in the alignment. GenBank
accession numbers given in parentheses are PH0574 (NP_142539),
mbAlaX (ZP_00296079), pfAlaRS (NP_577999), ecAlaRS (NP_417177),
saThrRS (NP_646443, PDB ID: 1NYR), ecThrRS (NP_416234, PDB ID:
1QF6).
|
 |
Figure 2.
Figure 2. (A) Ribbon diagram of PH0574, which consists of a
large (green) domain and a small (orange) domain. The Zn^2+ ion
is located between the two domains. (B) Close-up view of the
Zn^2+ ion binding site. A strong peak in the anomalous
difference Fourier map was observed only at the zinc position.
Many water molecules were observed around the Zn^2+ ion,
although none of these are involved in coordination. The
anomalous difference Fourier map contoured at 20  (orange)
and 2F[o] - F[c] map contoured at 2 (orange)
and 2F[o] - F[c] map contoured at 2  (blue)
are shown. (C) Zinc ion coordinate diagram. The coordination of
the Zn^2+ ion is mediated by the conserved histidine and
cysteine residues both in PH0574 (green) and in saThrRS (blue,
parentheses, PDB ID: 1NYR). (D) Structure of the putative active
site drawn with a van der Waals surface with charge. The cavity
accommodates an acyl serine. (blue)
are shown. (C) Zinc ion coordinate diagram. The coordination of
the Zn^2+ ion is mediated by the conserved histidine and
cysteine residues both in PH0574 (green) and in saThrRS (blue,
parentheses, PDB ID: 1NYR). (D) Structure of the putative active
site drawn with a van der Waals surface with charge. The cavity
accommodates an acyl serine.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2006,
62,
1133-1137)
copyright 2006.
|
|
| |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|

|
| |
The structural and functional report of the same protein, PH0574 as AlaX was published independently by Sogabe et al., and they confirmed the tRNA editing function experimentally.
Sokabe M, Okada A, Yao M, Nakashima T, Tanaka I (2005).
Molecular basis of alanine discrimination in editing site.
Proc. Natl. Acad. Sci. USA, 102, 11669-11674.
[PubMed: ]
PDB entries: and .
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Y.Mulkidjanian,
and
M.Y.Galperin
(2009).
On the origin of life in the Zinc world. 2. Validation of the hypothesis on the photosynthesizing zinc sulfide edifices as cradles of life on Earth.
|
| |
Biol Direct,
4,
27.
|
 |
|
|
|
|
 |
S.Kamijo,
A.Fujii,
K.Onodera,
and
K.Wakabayashi
(2009).
Analyses of conditions for KMSSS loop in tyrosyl-tRNA synthetase by building a mutant library.
|
| |
J Biochem,
146,
241-250.
|
 |
|
|
|
|
 |
B.Zhu,
M.W.Zhao,
G.Eriani,
and
E.D.Wang
(2007).
A present-day aminoacyl-tRNA synthetase with ancestral editing properties.
|
| |
RNA,
13,
15-21.
|
 |
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2007).
Structure of the AlaX-M trans-editing enzyme from Pyrococcus horikoshii.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
390-400.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2007).
Crystallization and preliminary X-ray crystallographic study of alanyl-tRNA synthetase from the archaeon Archaeoglobus fulgidus.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
224-228.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

