 |
PDBsum entry 1tkc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.2.1.1
- transketolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-sedoheptulose 7-phosphate + D-glyceraldehyde 3-phosphate = aldehydo-D- ribose 5-phosphate + D-xylulose 5-phosphate
|
 |
 |
 |
 |
 |
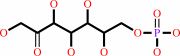 D-sedoheptulose 7-phosphate
D-sedoheptulose 7-phosphate
|
+
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
=
|
aldehydo-D- ribose 5-phosphate
|
+
|
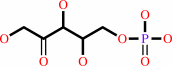 D-xylulose 5-phosphate
D-xylulose 5-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
M6T)
matches with 96.30% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biol Chem
269:10879-10882
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Specificity of coenzyme binding in thiamin diphosphate-dependent enzymes. Crystal structures of yeast transketolase in complex with analogs of thiamin diphosphate.
|
|
S.König,
A.Schellenberger,
H.Neef,
G.Schneider.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The three-dimensional structures of complexes of yeast apotransketolase with the
coenzyme analogs 6'-methyl, N1'-pyridyl, and N3'-pyridyl thiamin diphosphate,
respectively, were determined with protein crystallographic methods. All three
coenzyme analogs bind to the enzyme in a fashion highly similar to the cofactor
thiamin diphosphate. Thus, either one of the hydrogen bonds of the pyrimidine
ring nitrogens to the protein is sufficient for proper binding and positioning
of the cofactor. The lack of catalytic activity of the N3'-pyridyl analog is not
due to incorrect orientation of the pyrimidine ring, but results from the
absence of the hydrogen bond between the N1' nitrogen atom and the conserved
residue Glu418. The structure analysis provides further evidence for the
importance of this conserved interaction for enzymatic thiamin catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Agyei-Owusu,
and
F.J.Leeper
(2009).
Thiamin diphosphate in biological chemistry: analogues of thiamin diphosphate in studies of enzymes and riboswitches.
|
| |
FEBS J,
276,
2905-2916.
|
 |
|
|
|
|
 |
O.A.Esakova,
E.A.Khanova,
L.E.Meshalkina,
R.Golbik,
G.Hübner,
and
G.A.Kochetov
(2005).
Effect of transketolase substrates on holoenzyme reconstitution and stability.
|
| |
Biochemistry (Mosc),
70,
770-776.
|
 |
|
|
|
|
 |
R.Golbik,
L.E.Meshalkina,
T.Sandalova,
K.Tittmann,
E.Fiedler,
H.Neef,
S.König,
R.Kluger,
G.A.Kochetov,
G.Schneider,
and
G.Hübner
(2005).
Effect of coenzyme modification on the structural and catalytic properties of wild-type transketolase and of the variant E418A from Saccharomyces cerevisiae.
|
| |
FEBS J,
272,
1326-1342.
|
 |
|
|
|
|
 |
M.V.Kovina,
A.De Kok,
I.A.Sevostyanova,
L.S.Khailova,
N.V.Belkina,
and
G.A.Kochetov
(2004).
The molecular origin of the thiamine diphosphate-induced spectral bands of ThDP-dependent enzymes.
|
| |
Proteins,
56,
338-345.
|
 |
|
|
|
|
 |
O.A.Esakova,
L.E.Meshalkina,
R.Golbik,
G.Hübner,
and
G.A.Kochetov
(2004).
Donor substrate regulation of transketolase.
|
| |
Eur J Biochem,
271,
4189-4194.
|
 |
|
|
|
|
 |
G.Schenk,
R.G.Duggleby,
and
P.F.Nixon
(1998).
Properties and functions of the thiamin diphosphate dependent enzyme transketolase.
|
| |
Int J Biochem Cell Biol,
30,
1297-1318.
|
 |
|
|
|
|
 |
G.Schneider,
and
Y.Lindqvist
(1998).
Crystallography and mutagenesis of transketolase: mechanistic implications for enzymatic thiamin catalysis.
|
| |
Biochim Biophys Acta,
1385,
387-398.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

