 |
PDBsum entry 1thf

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.2.10
- imidazole glycerol-phosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-[(5-phospho-1-deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta- D-ribosyl)imidazole-4-carboxamide + L-glutamine = D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate + 5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide + L-glutamate + H+
|
 |
 |
 |
 |
 |
5-[(5-phospho-1-deoxy-D-ribulos-1-ylimino)methylamino]-1-(5-phospho-beta- D-ribosyl)imidazole-4-carboxamide
|
+
|
 L-glutamine
L-glutamine
|
=
|
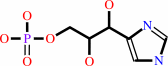 D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate
D-erythro-1-(imidazol- 4-yl)glycerol 3-phosphate
|
+
|
5-amino-1-(5-phospho-beta-D- ribosyl)imidazole-4-carboxamide
|
+
|
 L-glutamate
L-glutamate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
289:1546-1550
(2000)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion.
|
|
D.Lang,
R.Thoma,
M.Henn-Sax,
R.Sterner,
M.Wilmanns.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The atomic structures of two proteins in the histidine biosynthesis pathway
consist of beta/alpha barrels with a twofold repeat pattern. It is likely that
these proteins evolved by twofold gene duplication and gene fusion from a common
half-barrel ancestor. These ancestral domains are not visible as independent
domains in the extant proteins but can be inferred from a combination of
sequence and structural analysis. The detection of subdomain structures may be
useful in efforts to search genome sequences for functionally and structurally
related proteins.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Atomic structures of HisA (upper panel) and HisF (lower
panel) from Thermotoga maritima in ribbon presentations (29).
View from the COOH-terminal face of the central  barrel,
left; side view, center; and view from the NH[2]-terminal face
of the barrel, right. HisF contains two phosphate ions bound to
the active site, shown as space-filling models (red). The barrel,
left; side view, center; and view from the NH[2]-terminal face
of the barrel, right. HisF contains two phosphate ions bound to
the active site, shown as space-filling models (red). The  strands
and strands
and  helices
of the central eightfold helices
of the central eightfold  / /  barrel are
in orange and yellow, respectively. Loops at the NH[2]- and
COOH-terminal faces of the barrel are in cyan and green,
respectively. Some loops contain additional secondary structural
elements. The NH[2]- and COOH-termini are labeled when visible. barrel are
in orange and yellow, respectively. Loops at the NH[2]- and
COOH-terminal faces of the barrel are in cyan and green,
respectively. Some loops contain additional secondary structural
elements. The NH[2]- and COOH-termini are labeled when visible.
|
 |
Figure 3.
Fig. 3. Model for the evolution of the  / /  barrel
scaffold by twofold gene duplication. The first gene duplication
generates two initially identical half-barrels that are then
fused and adapted into an ancestral barrel
scaffold by twofold gene duplication. The first gene duplication
generates two initially identical half-barrels that are then
fused and adapted into an ancestral  / /  barrel. A
second gene duplication step leads to the diversification of the
ancestral barrel. A
second gene duplication step leads to the diversification of the
ancestral  / /  barrel
into two enzymes with distinct catalytic activities. barrel
into two enzymes with distinct catalytic activities.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(2000,
289,
1546-1550)
copyright 2000.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.V.Due,
J.Kuper,
A.Geerlof,
J.P.Kries,
and
M.Wilmanns
(2011).
Bisubstrate specificity in histidine/tryptophan biosynthesis isomerase from Mycobacterium tuberculosis by active site metamorphosis.
|
| |
Proc Natl Acad Sci U S A,
108,
3554-3559.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Setiyaputra,
J.P.Mackay,
and
W.M.Patrick
(2011).
The structure of a truncated phosphoribosylanthranilate isomerase suggests a unified model for evolution of the (βα)8 barrel fold.
|
| |
J Mol Biol,
408,
291-303.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Liebold,
F.List,
H.R.Kalbitzer,
R.Sterner,
and
E.Brunner
(2010).
The interaction of ammonia and xenon with the imidazole glycerol phosphate synthase from Thermotoga maritima as detected by NMR spectroscopy.
|
| |
Protein Sci,
19,
1774-1782.
|
 |
|
|
|
|
 |
C.Nagao,
N.Nagano,
and
K.Mizuguchi
(2010).
Relationships between functional subclasses and information contained in active-site and ligand-binding residues in diverse superfamilies.
|
| |
Proteins,
78,
2369-2384.
|
 |
|
|
|
|
 |
I.Yadid,
N.Kirshenbaum,
M.Sharon,
O.Dym,
and
D.S.Tawfik
(2010).
Metamorphic proteins mediate evolutionary transitions of structure.
|
| |
Proc Natl Acad Sci U S A,
107,
7287-7292.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Feng,
M.Li,
Y.Huang,
and
Y.Xiao
(2010).
Symmetric key structural residues in symmetric proteins with beta-trefoil fold.
|
| |
PLoS One,
5,
e14138.
|
 |
|
|
|
|
 |
S.Akanuma,
T.Matsuba,
E.Ueno,
N.Umeda,
and
A.Yamagishi
(2010).
Mimicking the evolution of a thermally stable monomeric four-helix bundle by fusion of four identical single-helix peptides.
|
| |
J Biochem,
147,
371-379.
|
 |
|
|
|
|
 |
S.Eisenbeis,
and
B.Höcker
(2010).
Evolutionary mechanism as a template for protein engineering.
|
| |
J Pept Sci,
16,
538-544.
|
 |
|
|
|
|
 |
A.Fischer,
N.Enkler,
G.Neudert,
M.Bocola,
R.Sterner,
and
R.Merkl
(2009).
TransCent: computational enzyme design by transferring active sites and considering constraints relevant for catalysis.
|
| |
BMC Bioinformatics,
10,
54.
|
 |
|
|
|
|
 |
H.Li,
W.Fast,
and
S.J.Benkovic
(2009).
Structural and functional modularity of proteins in the de novo purine biosynthetic pathway.
|
| |
Protein Sci,
18,
881-892.
|
 |
|
|
|
|
 |
J.Claren,
C.Malisi,
B.Höcker,
and
R.Sterner
(2009).
Establishing wild-type levels of catalytic activity on natural and artificial (beta alpha)8-barrel protein scaffolds.
|
| |
Proc Natl Acad Sci U S A,
106,
3704-3709.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Lipchock,
and
J.P.Loria
(2009).
Millisecond dynamics in the allosteric enzyme imidazole glycerol phosphate synthase (IGPS) from Thermotoga maritima.
|
| |
J Biomol NMR,
45,
73-84.
|
 |
|
|
|
|
 |
M.Hidaka,
M.Nishimoto,
M.Kitaoka,
T.Wakagi,
H.Shoun,
and
S.Fushinobu
(2009).
The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: a large deformation of a TIM barrel scaffold.
|
| |
J Biol Chem,
284,
7273-7283.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Biegert,
and
J.Söding
(2008).
De novo identification of highly diverged protein repeats by probabilistic consistency.
|
| |
Bioinformatics,
24,
807-814.
|
 |
|
|
|
|
 |
A.N.Alexandrova,
D.Röthlisberger,
D.Baker,
and
W.L.Jorgensen
(2008).
Catalytic mechanism and performance of computationally designed enzymes for Kemp elimination.
|
| |
J Am Chem Soc,
130,
15907-15915.
|
 |
|
|
|
|
 |
F.Birzele,
G.Csaba,
and
R.Zimmer
(2008).
Alternative splicing and protein structure evolution.
|
| |
Nucleic Acids Res,
36,
550-558.
|
 |
|
|
|
|
 |
I.Chaudhuri,
J.Söding,
and
A.N.Lupas
(2008).
Evolution of the beta-propeller fold.
|
| |
Proteins,
71,
795-803.
|
 |
|
|
|
|
 |
J.J.Graziano,
W.Liu,
R.Perera,
B.H.Geierstanger,
S.A.Lesley,
and
P.G.Schultz
(2008).
Selecting folded proteins from a library of secondary structural elements.
|
| |
J Am Chem Soc,
130,
176-185.
|
 |
|
|
|
|
 |
J.M.Lipchock,
and
J.P.Loria
(2008).
1H, 15N and 13C resonance assignment of imidazole glycerol phosphate (IGP) synthase protein HisF from Thermotoga maritima.
|
| |
Biomol NMR Assign,
2,
219-221.
|
 |
|
|
|
|
 |
K.K.Chan,
A.A.Fedorov,
E.V.Fedorov,
S.C.Almo,
and
J.A.Gerlt
(2008).
Structural basis for substrate specificity in phosphate binding (beta/alpha)8-barrels: D-allulose 6-phosphate 3-epimerase from Escherichia coli K-12.
|
| |
Biochemistry,
47,
9608-9617.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Li,
Y.Huang,
and
Y.Xiao
(2008).
Effects of external interactions on protein sequence-structure relations of beta-trefoil fold.
|
| |
Proteins,
72,
1161-1170.
|
 |
|
|
|
|
 |
M.T.Reetz,
M.Rentzsch,
A.Pletsch,
A.Taglieber,
F.Hollmann,
R.J.Mondière,
N.Dickmann,
B.Höcker,
S.Cerrone,
M.C.Haeger,
and
R.Sterner
(2008).
A robust protein host for anchoring chelating ligands and organocatalysts.
|
| |
Chembiochem,
9,
552-564.
|
 |
|
|
|
|
 |
T.A.Bharat,
S.Eisenbeis,
K.Zeth,
and
B.Höcker
(2008).
A beta alpha-barrel built by the combination of fragments from different folds.
|
| |
Proc Natl Acad Sci U S A,
105,
9942-9947.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Payandeh,
and
E.F.Pai
(2007).
Enzyme-driven speciation: crystallizing Archaea via lipid capture.
|
| |
J Mol Evol,
64,
364-374.
|
 |
|
|
|
|
 |
N.Lartillot,
H.Brinkmann,
and
H.Philippe
(2007).
Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model.
|
| |
BMC Evol Biol,
7,
S4.
|
 |
|
|
|
|
 |
P.L.Nester,
E.Gayó,
C.Latorre,
T.E.Jordan,
and
N.Blanco
(2007).
Perennial stream discharge in the hyperarid Atacama Desert of northern Chile during the latest Pleistocene.
|
| |
Proc Natl Acad Sci U S A,
104,
19724-19729.
|
 |
|
|
|
|
 |
R.Fani,
M.Brilli,
M.Fondi,
and
P.Lió
(2007).
The role of gene fusions in the evolution of metabolic pathways: the histidine biosynthesis case.
|
| |
BMC Evol Biol,
7,
S4.
|
 |
|
|
|
|
 |
T.Ke,
X.D.Ma,
P.H.Mao,
X.Jin,
S.J.Chen,
Y.Li,
L.X.Ma,
and
G.Y.He
(2007).
A mutant alpha-amylase with only part of the catalytic domain and its structural implication.
|
| |
Biotechnol Lett,
29,
117-122.
|
 |
|
|
|
|
 |
Y.Huang,
and
Y.Xiao
(2007).
Detection of gene duplication signals of Ig folds from their amino acid sequences.
|
| |
Proteins,
68,
267-272.
|
 |
|
|
|
|
 |
C.Mir,
E.Lopez-Viñas,
R.Aledo,
B.Puisac,
C.Rizzo,
C.Dionisi-Vici,
F.Deodato,
J.Pié,
P.Gomez-Puertas,
F.G.Hegardt,
and
N.Casals
(2006).
A single-residue mutation, G203E, causes 3-hydroxy-3-methylglutaric aciduria by occluding the substrate channel in the 3D structural model of HMG-CoA lyase.
|
| |
J Inherit Metab Dis,
29,
64-70.
|
 |
|
|
|
|
 |
J.Söding,
M.Remmert,
and
A.Biegert
(2006).
HHrep: de novo protein repeat detection and the origin of TIM barrels.
|
| |
Nucleic Acids Res,
34,
W137-W142.
|
 |
|
|
|
|
 |
S.Quevillon-Cheruel,
N.Leulliot,
M.Graille,
K.Blondeau,
J.Janin,
and
H.van Tilbeurgh
(2006).
Crystal structure of the yeast His6 enzyme suggests a reaction mechanism.
|
| |
Protein Sci,
15,
1516-1521.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Höcker
(2005).
Directed evolution of (betaalpha)(8)-barrel enzymes.
|
| |
Biomol Eng,
22,
31-38.
|
 |
|
|
|
|
 |
D.G.Naumoff
(2005).
GH97 is a new family of glycoside hydrolases, which is related to the alpha-galactosidase superfamily.
|
| |
BMC Genomics,
6,
112.
|
 |
|
|
|
|
 |
J.A.Gaspar,
C.Liu,
K.A.Vassall,
G.Meglei,
R.Stephen,
P.B.Stathopulos,
A.Pineda-Lucena,
B.Wu,
A.Yee,
C.H.Arrowsmith,
and
E.M.Meiering
(2005).
A novel member of the YchN-like fold: solution structure of the hypothetical protein Tm0979 from Thermotoga maritima.
|
| |
Protein Sci,
14,
216-223.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Kuper,
C.Doenges,
and
M.Wilmanns
(2005).
Two-fold repeated (betaalpha)4 half-barrels may provide a molecular tool for dual substrate specificity.
|
| |
EMBO Rep,
6,
134-139.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.C.Vega,
P.Zou,
F.J.Fernandez,
G.E.Murphy,
R.Sterner,
A.Popov,
and
M.Wilmanns
(2005).
Regulation of the hetero-octameric ATP phosphoribosyl transferase complex from Thermotoga maritima by a tRNA synthetase-like subunit.
|
| |
Mol Microbiol,
55,
675-686.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.E.Amaro,
R.S.Myers,
V.J.Davisson,
and
Z.A.Luthey-Schulten
(2005).
Structural elements in IGP synthase exclude water to optimize ammonia transfer.
|
| |
Biophys J,
89,
475-487.
|
 |
|
|
|
|
 |
V.K.Dubey,
J.Lee,
and
M.Blaber
(2005).
Redesigning symmetry-related "mini-core" regions of FGF-1 to increase primary structure symmetry: thermodynamic and functional consequences of structural symmetry.
|
| |
Protein Sci,
14,
2315-2323.
|
 |
|
|
|
|
 |
W.M.Patrick,
and
J.M.Blackburn
(2005).
In vitro selection and characterization of a stable subdomain of phosphoribosylanthranilate isomerase.
|
| |
FEBS J,
272,
3684-3697.
|
 |
|
|
|
|
 |
A.Shukla,
and
P.Guptasarma
(2004).
Folding of beta/alpha-unit scrambled forms of S. cerevisiae triosephosphate isomerase: Evidence for autonomy of substructure formation and plasticity of hydrophobic and hydrogen bonding interactions in core of (beta/alpha)8-barrel.
|
| |
Proteins,
55,
548-557.
|
 |
|
|
|
|
 |
B.Höcker,
J.Claren,
and
R.Sterner
(2004).
Mimicking enzyme evolution by generating new (betaalpha)8-barrels from (betaalpha)4-half-barrels.
|
| |
Proc Natl Acad Sci U S A,
101,
16448-16453.
|
 |
|
|
|
|
 |
H.Wright,
F.Barona-Gómez,
D.A.Hodgson,
and
V.Fülöp
(2004).
Expression, purification and preliminary crystallographic analysis of phosphoribosyl isomerase (PriA) from Streptomyces coelicolor.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
534-536.
|
 |
|
|
|
|
 |
K.B.Murray,
W.R.Taylor,
and
J.M.Thornton
(2004).
Toward the detection and validation of repeats in protein structure.
|
| |
Proteins,
57,
365-380.
|
 |
|
|
|
|
 |
K.J.Woycechowsky
(2004).
Recombination of fragmented proteins.
|
| |
Chem Biol,
11,
589-591.
|
 |
|
|
|
|
 |
M.H.Ali,
E.Peisach,
K.N.Allen,
and
B.Imperiali
(2004).
X-ray structure analysis of a designed oligomeric miniprotein reveals a discrete quaternary architecture.
|
| |
Proc Natl Acad Sci U S A,
101,
12183-12188.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.J.Bernett,
T.Somasundaram,
and
M.Blaber
(2004).
An atomic resolution structure for human fibroblast growth factor 1.
|
| |
Proteins,
57,
626-634.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Nicolet,
and
C.L.Drennan
(2004).
AdoMet radical proteins--from structure to evolution--alignment of divergent protein sequences reveals strong secondary structure element conservation.
|
| |
Nucleic Acids Res,
32,
4015-4025.
|
 |
|
|
|
|
 |
A.C.Joerger,
S.Mayer,
and
A.R.Fersht
(2003).
Mimicking natural evolution in vitro: an N-acetylneuraminate lyase mutant with an increased dihydrodipicolinate synthase activity.
|
| |
Proc Natl Acad Sci U S A,
100,
5694-5699.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Matte,
J.Sivaraman,
I.Ekiel,
K.Gehring,
Z.Jia,
and
M.Cygler
(2003).
Contribution of structural genomics to understanding the biology of Escherichia coli.
|
| |
J Bacteriol,
185,
3994-4002.
|
 |
|
|
|
|
 |
B.A.Manjasetty,
J.Powlowski,
and
A.Vrielink
(2003).
Crystal structure of a bifunctional aldolase-dehydrogenase: sequestering a reactive and volatile intermediate.
|
| |
Proc Natl Acad Sci U S A,
100,
6992-6997.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.N.Chaudhuri,
M.R.Sawaya,
C.Y.Kim,
G.S.Waldo,
M.S.Park,
T.C.Terwilliger,
and
T.O.Yeates
(2003).
The crystal structure of the first enzyme in the pantothenate biosynthetic pathway, ketopantoate hydroxymethyltransferase, from M tuberculosis.
|
| |
Structure,
11,
753-764.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
F.Barona-Gómez,
and
D.A.Hodgson
(2003).
Occurrence of a putative ancient-like isomerase involved in histidine and tryptophan biosynthesis.
|
| |
EMBO Rep,
4,
296-300.
|
 |
|
|
|
|
 |
M.Hartmann,
T.R.Schneider,
A.Pfeil,
G.Heinrich,
W.N.Lipscomb,
and
G.H.Braus
(2003).
Evolution of feedback-inhibited beta /alpha barrel isoenzymes by gene duplication and a single mutation.
|
| |
Proc Natl Acad Sci U S A,
100,
862-867.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.R.Brych,
J.Kim,
T.M.Logan,
and
M.Blaber
(2003).
Accommodation of a highly symmetric core within a symmetric protein superfold.
|
| |
Protein Sci,
12,
2704-2718.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Douangamath,
M.Walker,
S.Beismann-Driemeyer,
M.C.Vega-Fernandez,
R.Sterner,
and
M.Wilmanns
(2002).
Structural evidence for ammonia tunneling across the (beta alpha)(8) barrel of the imidazole glycerol phosphate synthase bienzyme complex.
|
| |
Structure,
10,
185-193.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.A.Barbosa,
J.Sivaraman,
Y.Li,
R.Larocque,
A.Matte,
J.D.Schrag,
and
M.Cygler
(2002).
Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
|
| |
Proc Natl Acad Sci U S A,
99,
1859-1864.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Mayans,
A.Ivens,
L.J.Nissen,
K.Kirschner,
and
M.Wilmanns
(2002).
Structural analysis of two enzymes catalysing reverse metabolic reactions implies common ancestry.
|
| |
EMBO J,
21,
3245-3254.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Korolev,
T.Skarina,
E.Evdokimova,
S.Beasley,
A.Edwards,
A.Joachimiak,
and
A.Savchenko
(2002).
Crystal structure of glutamine amidotransferase from Thermotoga maritima.
|
| |
Proteins,
49,
420-422.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Höcker,
C.Jürgens,
M.Wilmanns,
and
R.Sterner
(2001).
Stability, catalytic versatility and evolution of the (beta alpha)(8)-barrel fold.
|
| |
Curr Opin Biotechnol,
12,
376-381.
|
 |
|
|
|
|
 |
F.Forcellino,
and
P.Derreumaux
(2001).
Computer simulations aimed at structure prediction of supersecondary motifs in proteins.
|
| |
Proteins,
45,
159-166.
|
 |
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2001).
Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies.
|
| |
Annu Rev Biochem,
70,
209-246.
|
 |
|
|
|
|
 |
J.D.Stevenson,
S.Lutz,
and
S.J.Benkovic
(2001).
Retracing Enzyme Evolution in the (betaalpha)(8)-Barrel Scaffold.
|
| |
Angew Chem Int Ed Engl,
40,
1854-1856.
|
 |
|
|
|
|
 |
J.Jung,
and
B.Lee
(2001).
Circularly permuted proteins in the protein structure database.
|
| |
Protein Sci,
10,
1881-1886.
|
 |
|
|
|
|
 |
J.M.Petock,
I.Y.Torshin,
Y.F.Wang,
G.C.Du Bois,
C.M.Croce,
R.W.Harrison,
and
I.T.Weber
(2001).
Structure of murine Tcl1 at 2.5 A resolution and implications for the TCL oncogene family.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1545-1551.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Henn-Sax,
B.Höcker,
M.Wilmanns,
and
R.Sterner
(2001).
Divergent evolution of (betaalpha)8-barrel enzymes.
|
| |
Biol Chem,
382,
1315-1320.
|
 |
|
|
|
|
 |
M.J.Banfield,
J.S.Lott,
V.L.Arcus,
A.A.McCarthy,
and
E.N.Baker
(2001).
Structure of HisF, a histidine biosynthetic protein from Pyrobaculum aerophilum.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
1518-1525.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.J.Klem,
Y.Chen,
and
V.J.Davisson
(2001).
Subunit interactions and glutamine utilization by Escherichia coli imidazole glycerol phosphate synthase.
|
| |
J Bacteriol,
183,
989-996.
|
 |
|
|
|
|
 |
C.Binda,
R.T.Bossi,
S.Wakatsuki,
S.Arzt,
A.Coda,
B.Curti,
M.A.Vanoni,
and
A.Mattevi
(2000).
Cross-talk and ammonia channeling between active centers in the unexpected domain arrangement of glutamate synthase.
|
| |
Structure,
8,
1299-1308.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Gerlt,
and
P.C.Babbitt
(2000).
Can sequence determine function?
|
| |
Genome Biol,
1,
REVIEWS0005.
|
 |
|
|
|
|
 |
J.Stubbe,
and
L.N.Johnson
(2000).
Clarity through structures. Editorial overview.
|
| |
Curr Opin Struct Biol,
10,
709-710.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

