 |
PDBsum entry 1t6d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.6.1.11
- exopolyphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[phosphate](n) + H2O = [phosphate](n-1) + phosphate + H+
|
 |
 |
 |
 |
 |
[phosphate](n)
|
+
|
H2O
|
=
|
[phosphate](n-1)
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.6.1.40
- guanosine-5'-triphosphate,3'-diphosphate diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
guanosine 3'-diphosphate 5'-triphosphate + H2O = guanosine 3',5'-bis(diphosphate) + phosphate + H+
|
 |
 |
 |
 |
 |
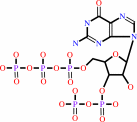 guanosine 3'-diphosphate 5'-triphosphate
guanosine 3'-diphosphate 5'-triphosphate
|
+
|
H2O
|
=
|
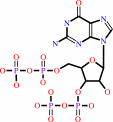 guanosine 3',5'-bis(diphosphate)
guanosine 3',5'-bis(diphosphate)
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
43:8894-8900
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural characterization of the stringent response related exopolyphosphatase/guanosine pentaphosphate phosphohydrolase protein family.
|
|
O.Kristensen,
M.Laurberg,
A.Liljas,
J.S.Kastrup,
M.Gajhede.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Exopolyphosphatase/guanosine pentaphosphate phosphohydrolase (PPX/GPPA) enzymes
play central roles in the bacterial stringent response induced by starvation.
The high-resolution crystal structure of the putative Aquifex aeolicus PPX/GPPA
phosphatase from the actin-like ATPase domain superfamily has been determined,
providing the first insights to features of the common catalytic core of the
PPX/GPPA family. The protein has a two-domain structure with an active site
located in the interdomain cleft. Two crystal forms were investigated (type I
and II) at resolutions of 1.53 and 2.15 A, respectively. This revealed a
structural flexibility that has previously been described as a "butterfly-like"
cleft opening around the active site in other actin-like superfamily proteins. A
calcium ion is observed at the center of this region in type I crystals,
substantiating that PPX/GPPA enzymes use metal ions for catalysis. Structural
analysis suggests that nucleotides bind at a similar position to that seen in
other members of the superfamily.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.F.Knowles
(2011).
The GDA1_CD39 superfamily: NTPDases with diverse functions.
|
| |
Purinergic Signal,
7,
21-45.
|
 |
|
|
|
|
 |
J.P.Vivian,
P.Riedmaier,
H.Ge,
J.Le Nours,
F.M.Sansom,
M.C.Wilce,
E.Byres,
M.Dias,
J.W.Schmidberger,
P.J.Cowan,
A.J.d'Apice,
E.L.Hartland,
J.Rossjohn,
and
T.Beddoe
(2010).
Crystal structure of a Legionella pneumophila ecto -triphosphate diphosphohydrolase, a structural and functional homolog of the eukaryotic NTPDases.
|
| |
Structure,
18,
228-238.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
N.N.Rao,
M.R.Gómez-García,
and
A.Kornberg
(2009).
Inorganic polyphosphate: essential for growth and survival.
|
| |
Annu Rev Biochem,
78,
605-647.
|
 |
|
|
|
|
 |
M.Zebisch,
and
N.Sträter
(2008).
Structural insight into signal conversion and inactivation by NTPDase2 in purinergic signaling.
|
| |
Proc Natl Acad Sci U S A,
105,
6882-6887.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Fang,
F.A.Ruiz,
M.Docampo,
S.Luo,
J.C.Rodrigues,
L.S.Motta,
P.Rohloff,
and
R.Docampo
(2007).
Overexpression of a Zn2+-sensitive soluble exopolyphosphatase from Trypanosoma cruzi depletes polyphosphate and affects osmoregulation.
|
| |
J Biol Chem,
282,
32501-32510.
|
 |
|
|
|
|
 |
M.Tammenkoski,
V.M.Moiseev,
M.Lahti,
E.Ugochukwu,
T.H.Brondijk,
S.A.White,
R.Lahti,
and
A.A.Baykov
(2007).
Kinetic and mutational analyses of the major cytosolic exopolyphosphatase from Saccharomyces cerevisiae.
|
| |
J Biol Chem,
282,
9302-9311.
|
 |
|
|
|
|
 |
J.Alvarado,
A.Ghosh,
T.Janovitz,
A.Jauregui,
M.S.Hasson,
and
D.A.Sanders
(2006).
Origin of exopolyphosphatase processivity: Fusion of an ASKHA phosphotransferase and a cyclic nucleotide phosphodiesterase homolog.
|
| |
Structure,
14,
1263-1272.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
T.L.Kirley,
P.A.Crawford,
and
T.M.Smith
(2006).
The structure of the nucleoside triphosphate diphosphohydrolases (NTPDases) as revealed by mutagenic and computational modeling analyses.
|
| |
Purinergic Signal,
2,
379-389.
|
 |
|
|
|
|
 |
Z.Liu,
and
R.A.Butow
(2006).
Mitochondrial retrograde signaling.
|
| |
Annu Rev Genet,
40,
159-185.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

