 |
PDBsum entry 1t5h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.2.1.33
- 4-chlorobenzoate--CoA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-chlorobenzoate + ATP + CoA = 4-chlorobenzoyl-CoA + AMP + diphosphate
|
 |
 |
 |
 |
 |
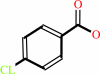 4-chlorobenzoate
4-chlorobenzoate
|
+
|
 ATP
ATP
|
+
|
 CoA
CoA
|
=
|
 4-chlorobenzoyl-CoA
4-chlorobenzoyl-CoA
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Mg(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
43:8670-8679
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of 4-chlorobenzoate:CoA ligase/synthetase in the unliganded and aryl substrate-bound states.
|
|
A.M.Gulick,
X.Lu,
D.Dunaway-Mariano.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
4-Chlorobenzoate:CoA ligase (CBAL) is a member of a family of adenylate-forming
enzymes that catalyze two-step adenylation and thioester-forming reactions. In
previous studies, we have provided structural evidence that members of this
enzyme family (exemplified by acetyl-CoA synthetase) use a large domain rotation
to catalyze the respective partial reactions [A. M. Gulick, V. J. Starai, A. R.
Horswill, K. M. Homick, and J. C. Escalante-Semerena, (2003) Biochemistry 42,
2866-2873]. CBAL catalyzes the synthesis of 4-chlorobenzoyl-CoA, the first step
in the 4-chlorobenzoate degredation pathway in PCB-degrading bacteria. We have
solved the 2.0 A crystal structure of the CBAL enzyme from Alcaligenes sp.
AL3007 using multiwavelength anomalous dispersion. The results demonstrate that
in the absence of any ligands, or bound to the aryl substrate 4-chlorobenzoate,
the enzyme adopts the conformation poised for catalysis of the adenylate-forming
half-reaction. We hypothesize that coenzyme A binding is required for
stabilization of the alternate conformation, which catalyzes the 4-CBA-CoA
thioester-forming reaction. We have also determined the structure of the enzyme
bound to the aryl substrate 4-chlorobenzoate. The aryl binding pocket is
composed of Phe184, His207, Val208, Val209, Phe249, Ala280, Ile303, Gly305,
Met310, and Asn311. The structure of the 4-chlorobenzoate binding site is
discussed in the context of the binding sites of other family members to gain
insight into substrate specificity and evolution of new function.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.J.Hughes,
and
A.Keatinge-Clay
(2011).
Enzymatic extender unit generation for in vitro polyketide synthase reactions: structural and functional showcasing of Streptomyces coelicolor MatB.
|
| |
Chem Biol,
18,
165-176.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Mao
(2011).
Dynamics studies of luciferase using elastic network model: how the sequence distribution of luciferase determines its color.
|
| |
Protein Eng Des Sel,
24,
341-349.
|
 |
|
|
|
|
 |
D.J.Sukovich,
J.L.Seffernick,
J.E.Richman,
J.A.Gralnick,
and
L.P.Wackett
(2010).
Widespread head-to-head hydrocarbon biosynthesis in bacteria and role of OleA.
|
| |
Appl Environ Microbiol,
76,
3850-3862.
|
 |
|
|
|
|
 |
H.Xu
(2010).
Enhancing MAD F(A) data for substructure determination.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
945-949.
|
 |
|
|
|
|
 |
T.V.Lee,
L.J.Johnson,
R.D.Johnson,
A.Koulman,
G.A.Lane,
J.S.Lott,
and
V.L.Arcus
(2010).
Structure of a eukaryotic nonribosomal peptide synthetase adenylation domain that activates a large hydroxamate amino acid in siderophore biosynthesis.
|
| |
J Biol Chem,
285,
2415-2427.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.M.Gulick
(2009).
Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase.
|
| |
ACS Chem Biol,
4,
811-827.
|
 |
|
|
|
|
 |
M.B.Shah,
C.Ingram-Smith,
L.L.Cooper,
J.Qu,
Y.Meng,
K.S.Smith,
and
A.M.Gulick
(2009).
The 2.1 A crystal structure of an acyl-CoA synthetase from Methanosarcina acetivorans reveals an alternate acyl-binding pocket for small branched acyl substrates.
|
| |
Proteins,
77,
685-698.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Wu,
A.S.Reger,
X.Lu,
A.M.Gulick,
and
D.Dunaway-Mariano
(2009).
The mechanism of domain alternation in the acyl-adenylate forming ligase superfamily member 4-chlorobenzoate: coenzyme A ligase.
|
| |
Biochemistry,
48,
4115-4125.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.S.Reger,
R.Wu,
D.Dunaway-Mariano,
and
A.M.Gulick
(2008).
Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
|
| |
Biochemistry,
47,
8016-8025.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Fraga
(2008).
Firefly luminescence: a historical perspective and recent developments.
|
| |
Photochem Photobiol Sci,
7,
146-158.
|
 |
|
|
|
|
 |
H.Xu,
and
C.M.Weeks
(2008).
Rapid and automated substructure solution by Shake-and-Bake.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
172-177.
|
 |
|
|
|
|
 |
H.Yonus,
P.Neumann,
S.Zimmermann,
J.J.May,
M.A.Marahiel,
and
M.T.Stubbs
(2008).
Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains.
|
| |
J Biol Chem,
283,
32484-32491.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.C.Chae,
B.Song,
and
G.J.Zylstra
(2008).
Identification of genes coding for hydrolytic dehalogenation in the metagenome derived from a denitrifying 4-chlorobenzoate degrading consortium.
|
| |
FEMS Microbiol Lett,
281,
203-209.
|
 |
|
|
|
|
 |
T.Abe,
Y.Hashimoto,
H.Hosaka,
K.Tomita-Yokotani,
and
M.Kobayashi
(2008).
Discovery of amide (peptide) bond synthetic activity in Acyl-CoA synthetase.
|
| |
J Biol Chem,
283,
11312-11321.
|
 |
|
|
|
|
 |
X.Lu,
H.Zhang,
P.J.Tonge,
and
D.S.Tan
(2008).
Mechanism-based inhibitors of MenE, an acyl-CoA synthetase involved in bacterial menaquinone biosynthesis.
|
| |
Bioorg Med Chem Lett,
18,
5963-5966.
|
 |
|
|
|
|
 |
A.S.Reger,
J.M.Carney,
and
A.M.Gulick
(2007).
Biochemical and crystallographic analysis of substrate binding and conformational changes in acetyl-CoA synthetase.
|
| |
Biochemistry,
46,
6536-6546.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Szarecka,
Y.Xu,
and
P.Tang
(2007).
Dynamics of firefly luciferase inhibition by general anesthetics: Gaussian and anisotropic network analyses.
|
| |
Biophys J,
93,
1895-1905.
|
 |
|
|
|
|
 |
E.Arias-Barrau,
E.R.Olivera,
A.Sandoval,
G.Naharro,
and
J.M.Luengo
(2006).
Acetyl-CoA synthetase from Pseudomonas putida U is the only acyl-CoA activating enzyme induced by acetate in this bacterium.
|
| |
FEMS Microbiol Lett,
260,
36-46.
|
 |
|
|
|
|
 |
E.J.Drake,
D.A.Nicolai,
and
A.M.Gulick
(2006).
Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain.
|
| |
Chem Biol,
13,
409-419.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.V.Somu,
D.J.Wilson,
E.M.Bennett,
H.I.Boshoff,
L.Celia,
B.J.Beck,
C.E.Barry,
and
C.C.Aldrich
(2006).
Antitubercular nucleosides that inhibit siderophore biosynthesis: SAR of the glycosyl domain.
|
| |
J Med Chem,
49,
7623-7635.
|
 |
|
|
|
|
 |
H.Xu,
C.M.Weeks,
and
H.A.Hauptman
(2005).
Optimizing statistical Shake-and-Bake for Se-atom substructure determination.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
976-981.
|
 |
|
|
|
|
 |
S.K.Samanta,
and
C.S.Harwood
(2005).
Use of the Rhodopseudomonas palustris genome sequence to identify a single amino acid that contributes to the activity of a coenzyme A ligase with chlorinated substrates.
|
| |
Mol Microbiol,
55,
1151-1159.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

