 |
PDBsum entry 1skb

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.1.3.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.8
- 3-phytase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1D-myo-inositol hexakisphosphate + H2O = 1D-myo-inositol 1,2,4,5,6- pentakisphosphate + phosphate
|
 |
 |
 |
 |
 |
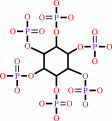 1D-myo-inositol hexakisphosphate
1D-myo-inositol hexakisphosphate
|
+
|
H2O
|
=
|
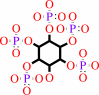 1D-myo-inositol 1,2,4,5,6- pentakisphosphate
1D-myo-inositol 1,2,4,5,6- pentakisphosphate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
12:1575-1583
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic snapshots of Aspergillus fumigatus phytase, revealing its enzymatic dynamics.
|
|
Q.Liu,
Q.Huang,
X.G.Lei,
Q.Hao.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Understanding of the atomic movements involved in an enzymatic reaction needs
structural information on the active and inactive native enzyme molecules and on
the enzyme-substrate, enzyme-intermediate, and enzyme-product(s) complexes. By
using the X-ray crystallographic method, four crystal structures of Aspergillus
fumigatus phytase were obtained at resolution higher than 1.7 A. The
pH-dependent catalytic activity of A. fumigatus phytase was linked to three
water molecules that may prevent the substrate from binding and thus block
nucleophilic attack of the catalytic imidazole nitrogen. Comparison of various
structures also identified the water molecule that attacks the phosphamide bond
during the hydrolysis process, and established the hydrolysis pathway of the
intermediate. Additionally, two reaction product phosphates were observed at the
active site, suggesting a possible product release pathway after hydrolysis of
the intermediate. These results can help explain the catalytic mechanism
throughout the whole acid phosphatase family, as all key residues are conserved.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 7.
Figure 7. The Release Pathway of Phosphate IonsPO4 501 is
the phosphate near the catalytic residue and PO4 502 is the
phosphate with hydrogen bonding to five water molecules. This
figure was prepared with Ligplot (Wallace et al., 1995).
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2004,
12,
1575-1583)
copyright 2004.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
W.Zhang,
and
X.G.Lei
(2008).
Cumulative improvements of thermostability and pH-activity profile of Aspergillus niger PhyA phytase by site-directed mutagenesis.
|
| |
Appl Microbiol Biotechnol,
77,
1033-1040.
|
 |
|
|
|
|
 |
W.Zhang,
E.J.Mullaney,
and
X.G.Lei
(2007).
Adopting selected hydrogen bonding and ionic interactions from Aspergillus fumigatus phytase structure improves the thermostability of Aspergillus niger PhyA phytase.
|
| |
Appl Environ Microbiol,
73,
3069-3076.
|
 |
|
|
|
|
 |
W.Morelle,
M.Bernard,
J.P.Debeaupuis,
M.Buitrago,
M.Tabouret,
and
J.P.Latgé
(2005).
Galactomannoproteins of Aspergillus fumigatus.
|
| |
Eukaryot Cell,
4,
1308-1316.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

