 |
PDBsum entry 1sd1

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase complexed with formycin a
|
|
Structure:
|
 |
5'-methylthioadenosine phosphorylase. Chain: a. Synonym: mta phosphorylase, mtapase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: mtap, msap. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Trimer (from PDB file)
Trimer (from PDB file)
|
|
Resolution:
|
 |
|
2.03Å
|
R-factor:
|
0.180
|
R-free:
|
0.198
|
|
|
Authors:
|
 |
J.E.Lee,E.C.Settembre,K.A.Cornell,M.K.Riscoe,J.R.Sufrin,S.E.Ealick, P.L.Howell
|
Key ref:

|
 |
J.E.Lee
et al.
(2004).
Structural comparison of MTA phosphorylase and MTA/AdoHcy nucleosidase explains substrate preferences and identifies regions exploitable for inhibitor design.
Biochemistry,
43,
5159-5169.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
12-Feb-04
|
Release date:
|
18-May-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q13126
(MTAP_HUMAN) -
S-methyl-5'-thioadenosine phosphorylase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
283 a.a.
268 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.28
- S-methyl-5'-thioadenosine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
S-methyl-5'-thioadenosine + phosphate = 5-(methylsulfanyl)-alpha-D-ribose 1-phosphate + adenine
|
 |
 |
 |
 |
 |
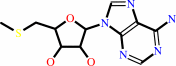 S-methyl-5'-thioadenosine
S-methyl-5'-thioadenosine
|
+
|
phosphate
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
=
|
5-(methylsulfanyl)-alpha-D-ribose 1-phosphate
|
+
|
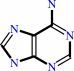 adenine
adenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
43:5159-5169
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural comparison of MTA phosphorylase and MTA/AdoHcy nucleosidase explains substrate preferences and identifies regions exploitable for inhibitor design.
|
|
J.E.Lee,
E.C.Settembre,
K.A.Cornell,
M.K.Riscoe,
J.R.Sufrin,
S.E.Ealick,
P.L.Howell.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The development of new and effective antiprotozoal drugs has been a difficult
challenge because of the close similarity of the metabolic pathways between
microbial and mammalian systems. 5'-Methylthioadenosine/S-adenosylhomocysteine
(MTA/AdoHcy) nucleosidase is thought to be an ideal target for therapeutic drug
design as the enzyme is present in many microbes but not in mammals. MTA/AdoHcy
nucleosidase (MTAN) irreversibly depurinates MTA or AdoHcy to form adenine and
the corresponding thioribose. The inhibition of MTAN leads to a buildup of toxic
byproducts that affect various microbial pathways such as quorum sensing,
biological methylation, polyamine biosynthesis, and methionine recycling. The
design of nucleosidase-specific inhibitors is complicated by its structural
similarity to the human MTA phosphorylase (MTAP). The crystal structures of
human MTAP complexed with formycin A and 5'-methylthiotubercidin have been
solved to 2.0 and 2.1 A resolution, respectively. Comparisons of the MTAP and
MTAN inhibitor complexes reveal size and electrostatic potential differences in
the purine, ribose, and 5'-alkylthio binding sites, which account for the
substrate specificity and reactions catalyzed. In addition, the differences
between the two enzymes have allowed the identification of exploitable regions
that can be targeted for the development of high-affinity nucleosidase-specific
inhibitors. Sequence alignments of Escherichia coli MTAN, human MTAP, and plant
MTA nucleosidases also reveal potential structural changes to the 5'-alkylthio
binding site that account for the substrate preference of plant MTA
nucleosidases.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.Parveen,
and
K.A.Cornell
(2011).
Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism.
|
| |
Mol Microbiol,
79,
7.
|
 |
|
|
|
|
 |
D.R.Ronning,
N.M.Iacopelli,
and
V.Mishra
(2010).
Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
|
| |
Protein Sci,
19,
2498-2510.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Albers
(2009).
Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5'-methylthioadenosine.
|
| |
IUBMB Life,
61,
1132-1142.
|
 |
|
|
|
|
 |
K.A.Cornell,
S.Primus,
J.A.Martinez,
and
N.Parveen
(2009).
Assessment of methylthioadenosine/S-adenosylhomocysteine nucleosidases of Borrelia burgdorferi as targets for novel antimicrobials using a novel high-throughput method.
|
| |
J Antimicrob Chemother,
63,
1163-1172.
|
 |
|
|
|
|
 |
E.Y.Park,
S.I.Oh,
M.J.Nam,
J.S.Shin,
K.N.Kim,
and
H.K.Song
(2006).
Crystal structure of 5'-methylthioadenosine nucleosidase from Arabidopsis thaliana at 1.5-A resolution.
|
| |
Proteins,
65,
519-523.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.L.Mosley,
B.A.Bakke,
J.M.Sadler,
N.K.Sunkara,
K.M.Dorgan,
Z.S.Zhou,
and
K.L.Seley-Radtke
(2006).
Carbocyclic pyrimidine nucleosides as inhibitors of S-adenosylhomocysteine hydrolase.
|
| |
Bioorg Med Chem,
14,
7967-7971.
|
 |
|
|
|
|
 |
A.Vendeville,
K.Winzer,
K.Heurlier,
C.M.Tang,
and
K.R.Hardie
(2005).
Making 'sense' of metabolism: autoinducer-2, LuxS and pathogenic bacteria.
|
| |
Nat Rev Microbiol,
3,
383-396.
|
 |
|
|
|
|
 |
E.Choi-Rhee,
and
J.E.Cronan
(2005).
A nucleosidase required for in vivo function of the S-adenosyl-L-methionine radical enzyme, biotin synthase.
|
| |
Chem Biol,
12,
589-593.
|
 |
|
|
|
|
 |
J.E.Lee,
V.Singh,
G.B.Evans,
P.C.Tyler,
R.H.Furneaux,
K.A.Cornell,
M.K.Riscoe,
V.L.Schramm,
and
P.L.Howell
(2005).
Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
|
| |
J Biol Chem,
280,
18274-18282.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.Singh,
G.B.Evans,
D.H.Lenz,
J.M.Mason,
K.Clinch,
S.Mee,
G.F.Painter,
P.C.Tyler,
R.H.Furneaux,
J.E.Lee,
P.L.Howell,
and
V.L.Schramm
(2005).
Femtomolar transition state analogue inhibitors of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli.
|
| |
J Biol Chem,
280,
18265-18273.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

