 |
PDBsum entry 1ral

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1ral
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.50
- 3alpha-hydroxysteroid 3-dehydrogenase (Si-specific).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a 3alpha-hydroxysteroid + NAD+ = a 3-oxosteroid + NADH + H+
|
|
2.
|
a 3alpha-hydroxysteroid + NADP+ = a 3-oxosteroid + NADPH + H+
|
|
 |
 |
 |
 |
 |
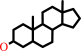 3alpha-hydroxysteroid
3alpha-hydroxysteroid
|
+
|
 NAD(+)
NAD(+)
|
=
|
 3-oxosteroid
3-oxosteroid
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 3alpha-hydroxysteroid
3alpha-hydroxysteroid
|
+
|
 NADP(+)
NADP(+)
|
=
|
 3-oxosteroid
3-oxosteroid
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
91:2517-2521
(1994)
|
|
PubMed id:
|
|
|
|

|
| |
|
Three-dimensional structure of rat liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase: a member of the aldo-keto reductase superfamily.
|
|
S.S.Hoog,
J.E.Pawlowski,
P.M.Alzari,
T.M.Penning,
M.Lewis.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The 3.0-A-resolution x-ray structure of rat liver 3 alpha-hydroxysteroid
dehydrogenase/dihydrodiol dehydrogenase (3 alpha-HSD, EC 1.1.1.50) was
determined by molecular replacement using human placental aldose reductase as
the search model. The protein folds into an alpha/beta or triose-phosphate
isomerase barrel and lacks a canonical Rossmann fold for binding pyridine
nucleotide. The structure contains a concentration of hydrophobic amino acids
that lie in a cavity near the top of the barrel and that are presumed to be
involved in binding hydrophobic substrates (steroids, prostaglandins, and
polycyclic aromatic hydrocarbons) and inhibitors (nonsteroidal antiinflammatory
drugs). At the distal end of this cavity lie three residues in close proximity
that have been implicated in catalysis by site-directed mutagenesis--Tyr-55,
Asp-50, and Lys-84. Tyr-55 is postulated to act as the general acid. 3 alpha-HSD
shares significant sequence identity with other HSDs that belong to the
aldo-keto reductase superfamily and these may show similar architecture. Other
members of this family include prostaglandin F synthase and rho-crystallin. By
contrast, 3 alpha-HSD shares no sequence identity with HSDs that are members of
the short-chain alcohol dehydrogenase family but does contain the
Tyr-Xaa-Xaa-Xaa-Lys consensus sequence implicated in catalysis in this family.
In the 3 alpha-HSD structure these residues are on the periphery of the barrel
and are unlikely to participate in catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
 Google scholar
Google scholar
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.Nagata,
Y.Kusakari,
Y.Fukunishi,
T.Inoue,
and
Y.Urade
(2011).
Catalytic mechanism of the primary human prostaglandin F2α synthase, aldo-keto reductase 1B1--prostaglandin D2 synthase activity in the absence of NADP(H).
|
| |
FEBS J,
278,
1288-1298.
|
 |
|
|
|
|
 |
J.L.Do Rego,
J.Y.Seong,
D.Burel,
J.Leprince,
V.Luu-The,
K.Tsutsui,
M.C.Tonon,
G.Pelletier,
and
H.Vaudry
(2009).
Neurosteroid biosynthesis: enzymatic pathways and neuroendocrine regulation by neurotransmitters and neuropeptides.
|
| |
Front Neuroendocrinol,
30,
259-301.
|
 |
|
|
|
|
 |
L.Di Costanzo,
J.E.Drury,
T.M.Penning,
and
D.W.Christianson
(2008).
Crystal structure of human liver Delta4-3-ketosteroid 5beta-reductase (AKR1D1) and implications for substrate binding and catalysis.
|
| |
J Biol Chem,
283,
16830-16839.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.Jiang,
C.Yang,
H.Qu,
Z.Liu,
Q.S.Fu,
and
C.Qiao
(2007).
Cloning of a novel aldo-keto reductase gene from Klebsiella sp. strain F51-1-2 and its functional expression in Escherichia coli.
|
| |
Appl Environ Microbiol,
73,
4959-4965.
|
 |
|
|
|
|
 |
U.Dhagat,
V.Carbone,
R.P.Chung,
C.Schulze-Briese,
S.Endo,
A.Hara,
and
O.El-Kabbani
(2007).
Structure of 3(17)alpha-hydroxysteroid dehydrogenase (AKR1C21) holoenzyme from an orthorhombic crystal form: an insight into the bifunctionality of the enzyme.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
825-830.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.C.Agís-Balboa,
G.Pinna,
A.Zhubi,
E.Maloku,
M.Veldic,
E.Costa,
and
A.Guidotti
(2006).
Characterization of brain neurons that express enzymes mediating neurosteroid biosynthesis.
|
| |
Proc Natl Acad Sci U S A,
103,
14602-14607.
|
 |
|
|
|
|
 |
Y.Jin,
and
T.M.Penning
(2006).
Multiple steps determine the overall rate of the reduction of 5alpha-dihydrotestosterone catalyzed by human type 3 3alpha-hydroxysteroid dehydrogenase: implications for the elimination of androgens.
|
| |
Biochemistry,
45,
13054-13063.
|
 |
|
|
|
|
 |
J.F.Couture,
K.P.de Jésus-Tran,
A.M.Roy,
L.Cantin,
P.L.Côté,
P.Legrand,
V.Luu-The,
F.Labrie,
and
R.Breton
(2005).
Comparison of crystal structures of human type 3 3alpha-hydroxysteroid dehydrogenase reveals an "induced-fit" mechanism and a conserved basic motif involved in the binding of androgen.
|
| |
Protein Sci,
14,
1485-1497.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Tsuruo
(2005).
Topography and function of androgen-metabolizing enzymes in the central nervous system.
|
| |
Anat Sci Int,
80,
1.
|
 |
|
|
|
|
 |
R.Takenoshita,
and
S.Toki
(2004).
[New aspects of hexobarbital metabolism: stereoselective metabolism, new metabolic pathway via GSH conjugation, and 3-hydroxyhexobarbital dehydrogenases]
|
| |
Yakugaku Zasshi,
124,
857-871.
|
 |
|
|
|
|
 |
T.M.Penning
(2004).
Aldo-keto reductases and formation of polycyclic aromatic hydrocarbon o-quinones.
|
| |
Methods Enzymol,
378,
31-67.
|
 |
|
|
|
|
 |
A.Ehrensberger,
and
D.K.Wilson
(2003).
Expression, crystallization and activities of the two family 11 aldo-keto reductases from Bacillus subtilis.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
375-377.
|
 |
|
|
|
|
 |
G.Obmolova,
A.Teplyakov,
P.P.Khil,
A.J.Howard,
R.D.Camerini-Otero,
and
G.L.Gilliland
(2003).
Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase.
|
| |
Proteins,
53,
323-325.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.W.Trauger,
A.Jiang,
B.A.Stearns,
and
P.V.LoGrasso
(2002).
Kinetics of allopregnanolone formation catalyzed by human 3 alpha-hydroxysteroid dehydrogenase type III (AKR1C2).
|
| |
Biochemistry,
41,
13451-13459.
|
 |
|
|
|
|
 |
M.L.Baker,
I.I.Serysheva,
S.Sencer,
Y.Wu,
S.J.Ludtke,
W.Jiang,
S.L.Hamilton,
and
W.Chiu
(2002).
The skeletal muscle Ca2+ release channel has an oxidoreductase-like domain.
|
| |
Proc Natl Acad Sci U S A,
99,
12155-12160.
|
 |
|
|
|
|
 |
B.Nidetzky,
P.Mayr,
W.Neuhauser,
and
M.Puchberger
(2001).
Structural and functional properties of aldose xylose reductase from the D-xylose-metabolizing yeast Candida tenuis.
|
| |
Chem Biol Interact,
130,
583-595.
|
 |
|
|
|
|
 |
R.Takenoshita,
Y.Nomura,
and
S.Toki
(2001).
Cloning and expression of cDNA encoding hamster liver 3-hydroxyhexobarbital/17beta(3alpha)-hydroxysteroid dehydrogenase 1.
|
| |
Chem Biol Interact,
130,
863-870.
|
 |
|
|
|
|
 |
E.Hur,
and
D.K.Wilson
(2000).
Crystallization and aldo-keto reductase activity of Gcy1p from Saccharomyces cerevisiae.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
763-765.
|
 |
|
|
|
|
 |
O.El-Kabbani,
H.Rogniaux,
P.Barth,
R.P.Chung,
E.V.Fletcher,
A.Van Dorsselaer,
and
A.Podjarny
(2000).
Aldose and aldehyde reductases: correlation of molecular modeling and mass spectrometric studies on the binding of inhibitors to the active site.
|
| |
Proteins,
41,
407-414.
|
 |
|
|
|
|
 |
Q.Ye,
D.Hyndman,
X.Li,
T.G.Flynn,
and
Z.Jia
(2000).
Crystal structure of CHO reductase, a member of the aldo-keto reductase superfamily.
|
| |
Proteins,
38,
41-48.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Khurana,
G.Sanli,
D.B.Powers,
S.Anderson,
and
M.Blaber
(2000).
Molecular modeling of substrate binding in wild-type and mutant Corynebacteria 2,5-diketo-D-gluconate reductases.
|
| |
Proteins,
39,
68-75.
|
 |
|
|
|
|
 |
H.Ma,
and
T.M.Penning
(1999).
Conversion of mammalian 3alpha-hydroxysteroid dehydrogenase to 20alpha-hydroxysteroid dehydrogenase using loop chimeras: changing specificity from androgens to progestins.
|
| |
Proc Natl Acad Sci U S A,
96,
11161-11166.
|
 |
|
|
|
|
 |
K.Ratnam,
H.Ma,
and
T.M.Penning
(1999).
The arginine 276 anchor for NADP(H) dictates fluorescence kinetic transients in 3 alpha-hydroxysteroid dehydrogenase, a representative aldo-keto reductase.
|
| |
Biochemistry,
38,
7856-7864.
|
 |
|
|
|
|
 |
B.P.Schlegel,
J.M.Jez,
and
T.M.Penning
(1998).
Mutagenesis of 3 alpha-hydroxysteroid dehydrogenase reveals a "push-pull" mechanism for proton transfer in aldo-keto reductases.
|
| |
Biochemistry,
37,
3538-3548.
|
 |
|
|
|
|
 |
B.P.Schlegel,
K.Ratnam,
and
T.M.Penning
(1998).
Retention of NADPH-linked quinone reductase activity in an aldo-keto reductase following mutation of the catalytic tyrosine.
|
| |
Biochemistry,
37,
11003-11011.
|
 |
|
|
|
|
 |
J.Luba,
B.Nare,
P.H.Liang,
K.S.Anderson,
S.M.Beverley,
and
L.W.Hardy
(1998).
Leishmania major pteridine reductase 1 belongs to the short chain dehydrogenase family: stereochemical and kinetic evidence.
|
| |
Biochemistry,
37,
4093-4104.
|
 |
|
|
|
|
 |
J.M.Jez,
and
T.M.Penning
(1998).
Engineering steroid 5 beta-reductase activity into rat liver 3 alpha-hydroxysteroid dehydrogenase.
|
| |
Biochemistry,
37,
9695-9703.
|
 |
|
|
|
|
 |
S.Khurana,
D.B.Powers,
S.Anderson,
and
M.Blaber
(1998).
Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution.
|
| |
Proc Natl Acad Sci U S A,
95,
6768-6773.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Bennett,
R.H.Albert,
J.M.Jez,
H.Ma,
T.M.Penning,
and
M.Lewis
(1997).
Steroid recognition and regulation of hormone action: crystal structure of testosterone and NADP+ bound to 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase.
|
| |
Structure,
5,
799-812.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.J.Liu,
Y.J.Sun,
J.Rose,
Y.J.Chung,
C.D.Hsiao,
W.R.Chang,
I.Kuo,
J.Perozich,
R.Lindahl,
J.Hempel,
and
B.C.Wang
(1997).
The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold.
|
| |
Nat Struct Biol,
4,
317-326.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Bennett,
B.P.Schlegel,
J.M.Jez,
T.M.Penning,
and
M.Lewis
(1996).
Structure of 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+.
|
| |
Biochemistry,
35,
10702-10711.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Janecek
(1996).
Invariant glycines and prolines flanking in loops the strand beta 2 of various (alpha/beta)8-barrel enzymes: a hidden homology?
|
| |
Protein Sci,
5,
1136-1143.
|
 |
|
|
|
|
 |
U.C.Oppermann,
and
E.Maser
(1996).
Characterization of a 3 alpha-hydroxysteroid dehydrogenase/carbonyl reductase from the gram-negative bacterium Comamonas testosteroni.
|
| |
Eur J Biochem,
241,
744-749.
|
 |
|
|
|
|
 |
W.L.Duax,
J.F.Griffin,
and
D.Ghosh
(1996).
The fascinating complexities of steroid-binding enzymes.
|
| |
Curr Opin Struct Biol,
6,
813-823.
|
 |
|
|
|
|
 |
S.W.Chouinard,
G.F.Wilson,
A.K.Schlimgen,
and
B.Ganetzky
(1995).
A potassium channel beta subunit related to the aldo-keto reductase superfamily is encoded by the Drosophila hyperkinetic locus.
|
| |
Proc Natl Acad Sci U S A,
92,
6763-6767.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

