 |
PDBsum entry 1qu9

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics
|
PDB id
|
|

|

|
1qu9
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.99.10
- 2-iminobutanoate/2-iminopropanoate deaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
2-iminobutanoate + H2O = 2-oxobutanoate + NH4+
|
|
2.
|
2-iminopropanoate + H2O = pyruvate + NH4+
|
|
 |
 |
 |
 |
 |
2-iminobutanoate
|
+
|
H2O
|
=
|
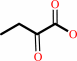 2-oxobutanoate
2-oxobutanoate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
2-iminopropanoate
|
+
|
H2O
|
=
|
 pyruvate
pyruvate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
8:2428-2437
(1999)
|
|
PubMed id:
|
|
|
|

|
| |
|
A test case for structure-based functional assignment: the 1.2 A crystal structure of the yjgF gene product from Escherichia coli.
|
|
K.Volz.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The YER057c/YIL051c/YjgF protein family is a set of 24 full-length homologs,
each approximately 130 residues in length, and each with no known function or
relationship to proteins of known structure. To determine the function of this
family, the structure of one member--the YjgF protein from Escherichia coli--was
solved and refined at a resolution of 1.2 A. The YjgF molecule is a homotrimer
with exact threefold symmetry. Its tertiary and quaternary structures are
related to that of Bacillus subtilis chorismate mutase, although their active
sites are completely different. The YjgF protein has an active site curiously
similar to protein tyrosine phosphatases, including a covalently modified
cysteine, but it is unlikely to be functionally related. The lessons learned
from this attempt to deduce function from structure may be useful to future
projects in structural genomics.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Karaca,
A.S.Melquiond,
S.J.de Vries,
P.L.Kastritis,
and
A.M.Bonvin
(2010).
Building macromolecular assemblies by information-driven docking: introducing the HADDOCK multibody docking server.
|
| |
Mol Cell Proteomics,
9,
1784-1794.
|
 |
|
|
|
|
 |
K.G.Thakur,
T.Praveena,
and
B.Gopal
(2010).
Mycobacterium tuberculosis Rv2704 is a member of the YjgF/YER057c/UK114 family.
|
| |
Proteins,
78,
773-778.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.J.Rangan,
Y.Y.Yang,
G.Charron,
and
H.C.Hang
(2010).
Rapid visualization and large-scale profiling of bacterial lipoproteins with chemical reporters.
|
| |
J Am Chem Soc,
132,
10628-10629.
|
 |
|
|
|
|
 |
K.S.Kim,
J.G.Pelton,
W.B.Inwood,
U.Andersen,
S.Kustu,
and
D.E.Wemmer
(2010).
The Rut pathway for pyrimidine degradation: novel chemistry and toxicity problems.
|
| |
J Bacteriol,
192,
4089-4102.
|
 |
|
|
|
|
 |
M.R.Christopherson,
G.E.Schmitz,
and
D.M.Downs
(2008).
YjgF is required for isoleucine biosynthesis when Salmonella enterica is grown on pyruvate medium.
|
| |
J Bacteriol,
190,
3057-3062.
|
 |
|
|
|
|
 |
N.J.Zelyas,
H.Cai,
T.Kwong,
and
S.E.Jensen
(2008).
Alanylclavam biosynthetic genes are clustered together with one group of clavulanic acid biosynthetic genes in Streptomyces clavuligerus.
|
| |
J Bacteriol,
190,
7957-7965.
|
 |
|
|
|
|
 |
J.D.Burman,
C.E.Stevenson,
R.G.Sawers,
and
D.M.Lawson
(2007).
The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family.
|
| |
BMC Struct Biol,
7,
30.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.A.Browne,
A.I.Ramos,
and
D.M.Downs
(2006).
PurF-independent phosphoribosyl amine formation in yjgF mutants of Salmonella enterica utilizes the tryptophan biosynthetic enzyme complex anthranilate synthase-phosphoribosyltransferase.
|
| |
J Bacteriol,
188,
6786-6792.
|
 |
|
|
|
|
 |
D.M.Downs
(2006).
Understanding microbial metabolism.
|
| |
Annu Rev Microbiol,
60,
533-559.
|
 |
|
|
|
|
 |
T.Miyakawa,
W.C.Lee,
K.Hatano,
Y.Kato,
Y.Sawano,
K.Miyazono,
K.Nagata,
and
M.Tanokura
(2006).
Crystal structure of the YjgF/YER057c/UK114 family protein from the hyperthermophilic archaeon Sulfolobus tokodaii strain 7.
|
| |
Proteins,
62,
557-561.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.Leitner-Dagan,
M.Ovadis,
A.Zuker,
E.Shklarman,
I.Ohad,
T.Tzfira,
and
A.Vainstein
(2006).
CHRD, a plant member of the evolutionarily conserved YjgF family, influences photosynthesis and chromoplastogenesis.
|
| |
Planta,
225,
89.
|
 |
|
|
|
|
 |
B.A.Manjasetty,
H.Delbrück,
D.T.Pham,
U.Mueller,
M.Fieber-Erdmann,
C.Scheich,
V.Sievert,
K.Büssow,
F.H.Niesen,
W.Weihofen,
B.Loll,
W.Saenger,
U.Heinemann,
and
F.H.Neisen
(2004).
Crystal structure of Homo sapiens protein hp14.5.
|
| |
Proteins,
54,
797-800.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Kolker,
K.S.Makarova,
S.Shabalina,
A.F.Picone,
S.Purvine,
T.Holzman,
T.Cherny,
D.Armbruster,
R.S.Munson,
G.Kolesov,
D.Frishman,
and
M.Y.Galperin
(2004).
Identification and functional analysis of 'hypothetical' genes expressed in Haemophilus influenzae.
|
| |
Nucleic Acids Res,
32,
2353-2361.
|
 |
|
|
|
|
 |
G.Schmitz,
and
D.M.Downs
(2004).
Reduced transaminase B (IlvE) activity caused by the lack of yjgF is dependent on the status of threonine deaminase (IlvA) in Salmonella enterica serovar Typhimurium.
|
| |
J Bacteriol,
186,
803-810.
|
 |
|
|
|
|
 |
V.Anantharaman,
and
L.Aravind
(2004).
The SHS2 module is a common structural theme in functionally diverse protein groups, like Rpb7p, FtsA, GyrI, and MTH1598/TM1083 superfamilies.
|
| |
Proteins,
56,
795-807.
|
 |
|
|
|
|
 |
A.Stark,
and
R.B.Russell
(2003).
Annotation in three dimensions. PINTS: Patterns in Non-homologous Tertiary Structures.
|
| |
Nucleic Acids Res,
31,
3341-3344.
|
 |
|
|
|
|
 |
D.Deriu,
C.Briand,
E.Mistiniene,
V.Naktinis,
and
M.G.Grütter
(2003).
Structure and oligomeric state of the mammalian tumour-associated antigen UK114.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1676-1678.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.Kolker,
S.Purvine,
M.Y.Galperin,
S.Stolyar,
D.R.Goodlett,
A.I.Nesvizhskii,
A.Keller,
T.Xie,
J.K.Eng,
E.Yi,
L.Hood,
A.F.Picone,
T.Cherny,
B.C.Tjaden,
A.F.Siegel,
T.J.Reilly,
K.S.Makarova,
B.O.Palsson,
and
A.L.Smith
(2003).
Initial proteome analysis of model microorganism Haemophilus influenzae strain Rd KW20.
|
| |
J Bacteriol,
185,
4593-4602.
|
 |
|
|
|
|
 |
J.D.Burman,
C.E.Stevenson,
K.A.Hauton,
G.Sawers,
and
D.M.Lawson
(2003).
Crystallization and preliminary X-ray analysis of the E. coli hypothetical protein TdcF.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1076-1078.
|
 |
|
|
|
|
 |
A.M.Deaconescu,
A.Roll-Mecak,
J.B.Bonanno,
S.E.Gerchman,
H.Kycia,
F.W.Studier,
and
S.K.Burley
(2002).
X-ray structure of Saccharomyces cerevisiae homologous mitochondrial matrix factor 1 (Hmf1).
|
| |
Proteins,
48,
431-436.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Lehmann,
K.Lim,
J.Toedt,
W.Krajewski,
A.Howard,
E.Eisenstein,
and
O.Herzberg
(2002).
Structure of 2C-methyl-D-erythrol-2,4-cyclodiphosphate synthase from Haemophilus influenzae: activation by conformational transition.
|
| |
Proteins,
49,
135-138.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.E.Kemp,
C.S.Bond,
and
W.N.Hunter
(2002).
Structure of 2C-methyl-D-erythritol 2,4- cyclodiphosphate synthase: an essential enzyme for isoprenoid biosynthesis and target for antimicrobial drug development.
|
| |
Proc Natl Acad Sci U S A,
99,
6591-6596.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Irving,
J.C.Whisstock,
and
A.M.Lesk
(2001).
Protein structural alignments and functional genomics.
|
| |
Proteins,
42,
378-382.
|
 |
|
|
|
|
 |
J.M.Kim,
H.Yoshikawa,
and
K.Shirahige
(2001).
A member of the YER057c/yjgf/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae.
|
| |
Genes Cells,
6,
507-517.
|
 |
|
|
|
|
 |
Z.Dauter,
M.Li,
and
A.Wlodawer
(2001).
Practical experience with the use of halides for phasing macromolecular structures: a powerful tool for structural genomics.
|
| |
Acta Crystallogr D Biol Crystallogr,
57,
239-249.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.H.Downing
(2000).
Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics.
|
| |
Annu Rev Cell Dev Biol,
16,
89.
|
 |
|
|
|
|
 |
M.Teplova,
V.Tereshko,
R.Sanishvili,
A.Joachimiak,
T.Bushueva,
W.F.Anderson,
and
M.Egli
(2000).
The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding.
|
| |
Protein Sci,
9,
2557-2566.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

