 |
PDBsum entry 1qap

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Glycosyltransferase
|
PDB id
|
|

|

|
1qap
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Glycosyltransferase
|
 |
|
Title:
|
 |
Quinolinic acid phosphoribosyltransferase with bound quinolinic acid
|
|
Structure:
|
 |
Quinolinic acid phosphoribosyltransferase. Chain: a, b. Synonym: quinolinate prtase, qaprtase, qaprt, qprt. Engineered: yes
|
|
Source:
|
 |
Salmonella typhimurium. Organism_taxid: 602. Cell_line: bl21. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.80Å
|
R-factor:
|
0.186
|
R-free:
|
0.274
|
|
|
Authors:
|
 |
J.C.Eads,D.Ozturk,T.B.Wexler,C.Grubmeyer,J.C.Sacchettini
|
Key ref:

|
 |
J.C.Eads
et al.
(1997).
A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase.
Structure,
5,
47-58.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
20-Sep-96
|
Release date:
|
12-Mar-97
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P30012
(NADC_SALTY) -
Nicotinate-nucleotide pyrophosphorylase [carboxylating] from Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
297 a.a.
289 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.19
- nicotinate-nucleotide diphosphorylase (carboxylating).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
nicotinate beta-D-ribonucleotide + CO2 + diphosphate = quinolinate + 5-phospho-alpha-D-ribose 1-diphosphate + 2 H+
|
 |
 |
 |
 |
 |
nicotinate beta-D-ribonucleotide
|
+
|
 CO2
CO2
|
+
|
 diphosphate
diphosphate
|
=
|
 quinolinate
quinolinate
|
+
|
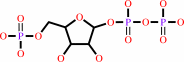 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
+
|
2
×
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
5:47-58
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
A new function for a common fold: the crystal structure of quinolinic acid phosphoribosyltransferase.
|
|
J.C.Eads,
D.Ozturk,
T.B.Wexler,
C.Grubmeyer,
J.C.Sacchettini.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
BACKGROUND: Quinolinic acid (QA) is a neurotoxin and has been shown to be
present at high levels in the central nervous system of patients with certain
diseases, such as AIDS and meningitis. The enzyme quinolinic acid
phosphoribosyltransferase (QAPRTase) provides the only route for QA metabolism
and is also an essential step in de novo NAD biosynthesis. QAPRTase catalyzes
the synthesis of nicotinic acid mononucleotide (NAMN) from QA and
5-phosphoribosyl-1-pyrophosphate (PRPP). The structures of several
phosphoribosyltransferases (PRTases) have been reported, and all have shown a
similar fold of a five-strandard beta sheet surrounded by four alpha helices. A
conserved sequence motif of 13 residues is common to these 'type I' PRTases but
is not observed in the QAPRTase sequence, suggestive of a different fold for
this enzyme. RESULTS: The crystal structure of QAPRTase from Salmonella
typhimurium has been determined with bound QA to 2.8 A resolution, and with
bound NAMN to 3.0 A resolution. Most significantly, the enzyme shows a
completely novel fold for a PRTase enzyme comprising a two-domain structure: a
mixed alpha/beta N-terminal domain and an alpha/beta barrel-like domain
containing seven beta strands. The active site is located at the C-terminal ends
of the beta strands of the alpha/beta barrel, and is bordered by the N-terminal
domain of the second subunit of the dimer. The active site is largely composed
of a number of conserved charged residues that appear to be important for
substrate binding and catalysis. CONCLUSIONS: The seven-stranded
alpha/beta-barrel domain of QAPRTase is very similar in structure to the
eight-stranded alpha/beta-barrel enzymes. The structure shows a
phosphate-binding site that appears to be conserved among many alpha/beta-barrel
enzymes including indole-3-glycerol phosphate synthase and flavocytochrome b2.
The new fold observed here demonstrates that the PRTase enzymes have evolved
their similar chemistry from at least two completely different protein
architectures.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 9.
Figure 9. Overlay of the phosphate-binding site regions of
QAPRTase (in blue), flavocytochrome b2 (in magenta) and
indole-3-glycerol phosphate synthase (in green). The bound NAMN
is shown in stick form with atom coloring as in Figure 6.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(1997,
5,
47-58)
copyright 1997.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Z.Bello,
B.Stitt,
and
C.Grubmeyer
(2010).
Interactions at the 2 and 5 positions of 5-phosphoribosyl pyrophosphate are essential in Salmonella typhimurium quinolinate phosphoribosyltransferase.
|
| |
Biochemistry,
49,
1377-1387.
|
 |
|
|
|
|
 |
Z.Bello,
and
C.Grubmeyer
(2010).
Roles for cationic residues at the quinolinic acid binding site of quinolinate phosphoribosyltransferase.
|
| |
Biochemistry,
49,
1388-1395.
|
 |
|
|
|
|
 |
M.K.Kim,
G.B.Kang,
W.K.Song,
and
S.H.Eom
(2007).
The role of Phe181 in the hexamerization of Helicobacter pylori quinolinate phosphoribosyltransferase.
|
| |
Protein J,
26,
517-521.
|
 |
|
|
|
|
 |
P.S.Monzani,
S.Trapani,
O.H.Thiemann,
and
G.Oliva
(2007).
Crystal structure of Leishmania tarentolae hypoxanthine-guanine phosphoribosyltransferase.
|
| |
BMC Struct Biol,
7,
59.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.A.Khan,
X.Tao,
and
L.Tong
(2006).
Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents.
|
| |
Nat Struct Mol Biol,
13,
582-588.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.K.Kim,
Y.J.Im,
J.H.Lee,
and
S.H.Eom
(2006).
Crystal structure of quinolinic acid phosphoribosyltransferase from Helicobacter pylori.
|
| |
Proteins,
63,
252-255.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Marino,
M.Deuss,
D.I.Svergun,
P.V.Konarev,
R.Sterner,
and
O.Mayans
(2006).
Structural and mutational analysis of substrate complexation by anthranilate phosphoribosyltransferase from Sulfolobus solfataricus.
|
| |
J Biol Chem,
281,
21410-21421.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Wang,
X.Zhang,
P.Bheda,
J.R.Revollo,
S.Imai,
and
C.Wolberger
(2006).
Structure of Nampt/PBEF/visfatin, a mammalian NAD+ biosynthetic enzyme.
|
| |
Nat Struct Mol Biol,
13,
661-662.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.H.Shin,
N.Oganesyan,
J.Jancarik,
H.Yokota,
R.Kim,
and
S.H.Kim
(2005).
Crystal structure of a nicotinate phosphoribosyltransferase from Thermoplasma acidophilum.
|
| |
J Biol Chem,
280,
18326-18335.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.S.Chappie,
J.M.Cànaves,
G.W.Han,
C.L.Rife,
Q.Xu,
and
R.C.Stevens
(2005).
The structure of a eukaryotic nicotinic acid phosphoribosyltransferase reveals structural heterogeneity among type II PRTases.
|
| |
Structure,
13,
1385-1396.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.S.Champagne,
M.Sissler,
Y.Larrabee,
S.Doublié,
and
C.S.Francklyn
(2005).
Activation of the hetero-octameric ATP phosphoribosyl transferase through subunit interface rearrangement by a tRNA synthetase paralog.
|
| |
J Biol Chem,
280,
34096-34104.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.C.Vega,
P.Zou,
F.J.Fernandez,
G.E.Murphy,
R.Sterner,
A.Popov,
and
M.Wilmanns
(2005).
Regulation of the hetero-octameric ATP phosphoribosyl transferase complex from Thermotoga maritima by a tRNA synthetase-like subunit.
|
| |
Mol Microbiol,
55,
675-686.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Kukimoto-Niino,
R.Shibata,
K.Murayama,
H.Hamana,
M.Nishimoto,
Y.Bessho,
T.Terada,
M.Shirouzu,
S.Kuramitsu,
and
S.Yokoyama
(2005).
Crystal structure of a predicted phosphoribosyltransferase (TT1426) from Thermus thermophilus HB8 at 2.01 A resolution.
|
| |
Protein Sci,
14,
823-827.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.Schwarzenbacher,
L.Jaroszewski,
F.von Delft,
P.Abdubek,
E.Ambing,
T.Biorac,
L.S.Brinen,
J.M.Canaves,
J.Cambell,
H.J.Chiu,
X.Dai,
A.M.Deacon,
M.DiDonato,
M.A.Elsliger,
S.Eshagi,
R.Floyd,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
E.Hampton,
C.Karlak,
H.E.Klock,
E.Koesema,
J.S.Kovarik,
A.Kreusch,
P.Kuhn,
S.A.Lesley,
I.Levin,
D.McMullan,
T.M.McPhillips,
M.D.Miller,
A.Morse,
K.Moy,
J.Ouyang,
R.Page,
K.Quijano,
A.Robb,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
X.Wang,
B.West,
G.Wolf,
Q.Xu,
K.O.Hodgson,
J.Wooley,
and
I.A.Wilson
(2004).
Crystal structure of a type II quinolic acid phosphoribosyltransferase (TM1645) from Thermotoga maritima at 2.50 A resolution.
|
| |
Proteins,
55,
768-771.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.V.Smith,
and
J.C.Sacchettini
(2003).
Mycobacterium tuberculosis: a model system for structural genomics.
|
| |
Curr Opin Struct Biol,
13,
658-664.
|
 |
|
|
|
|
 |
G.K.Grabner,
and
R.L.Switzer
(2003).
Kinetic studies of the uracil phosphoribosyltransferase reaction catalyzed by the Bacillus subtilis pyrimidine attenuation regulatory protein PyrR.
|
| |
J Biol Chem,
278,
6921-6927.
|
 |
|
|
|
|
 |
M.K.Kim,
Y.S.Kim,
S.H.Rho,
Y.J.Im,
J.H.Lee,
G.B.Kang,
and
S.H.Eom
(2003).
Crystallization and preliminary X-ray crystallographic analysis of quinolinate phosphoribosyltransferase of Helicobacter pylori.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1265-1266.
|
 |
|
|
|
|
 |
A.Kadziola,
J.Neuhard,
and
S.Larsen
(2002).
Structure of product-bound Bacillus caldolyticus uracil phosphoribosyltransferase confirms ordered sequential substrate binding.
|
| |
Acta Crystallogr D Biol Crystallogr,
58,
936-945.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Munagala,
V.J.Basus,
and
C.C.Wang
(2001).
Role of the flexible loop of hypoxanthine-guanine-xanthine phosphoribosyltransferase from Tritrichomonas foetus in enzyme catalysis.
|
| |
Biochemistry,
40,
4303-4311.
|
 |
|
|
|
|
 |
I.D'Angelo,
N.Raffaelli,
V.Dabusti,
T.Lorenzi,
G.Magni,
and
M.Rizzi
(2000).
Structure of nicotinamide mononucleotide adenylyltransferase: a key enzyme in NAD(+) biosynthesis.
|
| |
Structure,
8,
993.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Marković-Housley,
G.Miglierini,
L.Soldatova,
P.J.Rizkallah,
U.Müller,
and
T.Schirmer
(2000).
Crystal structure of hyaluronidase, a major allergen of bee venom.
|
| |
Structure,
8,
1025-1035.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Mattevi,
G.Tedeschi,
L.Bacchella,
A.Coda,
A.Negri,
and
S.Ronchi
(1999).
Structure of L-aspartate oxidase: implications for the succinate dehydrogenase/fumarate reductase oxidoreductase family.
|
| |
Structure,
7,
745-756.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.Lundegaard,
and
K.F.Jensen
(1999).
Kinetic mechanism of uracil phosphoribosyltransferase from Escherichia coli and catalytic importance of the conserved proline in the PRPP binding site.
|
| |
Biochemistry,
38,
3327-3334.
|
 |
|
|
|
|
 |
G.P.Wang,
C.Lundegaard,
K.F.Jensen,
and
C.Grubmeyer
(1999).
Kinetic mechanism of OMP synthase: a slow physical step following group transfer limits catalytic rate.
|
| |
Biochemistry,
38,
275-283.
|
 |
|
|
|
|
 |
L.Bacchella,
C.Lina,
F.Todone,
A.Negri,
G.Tedeschi,
S.Ronchi,
and
A.Mattevi
(1999).
Crystallization of L-aspartate oxidase, the first enzyme in the bacterial de novo biosynthesis of NAD.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
549-551.
|
 |
|
|
|
|
 |
C.C.Lee,
S.P.Craig,
and
A.E.Eakin
(1998).
A single amino acid substitution in the human and a bacterial hypoxanthine phosphoribosyltransferase modulates specificity for the binding of guanine.
|
| |
Biochemistry,
37,
3491-3498.
|
 |
|
|
|
|
 |
D.R.Tomchick,
R.J.Turner,
R.L.Switzer,
and
J.L.Smith
(1998).
Adaptation of an enzyme to regulatory function: structure of Bacillus subtilis PyrR, a pyr RNA-binding attenuation protein and uracil phosphoribosyltransferase.
|
| |
Structure,
6,
337-350.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.A.Schumacher,
D.Carter,
D.M.Scott,
D.S.Roos,
B.Ullman,
and
R.G.Brennan
(1998).
Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
|
| |
EMBO J,
17,
3219-3232.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Rajavel,
D.Lalo,
J.W.Gross,
and
C.Grubmeyer
(1998).
Conversion of a cosubstrate to an inhibitor: phosphorylation mutants of nicotinic acid phosphoribosyltransferase.
|
| |
Biochemistry,
37,
4181-4188.
|
 |
|
|
|
|
 |
P.J.Focia,
S.P.Craig,
and
A.E.Eakin
(1998).
Approaching the transition state in the crystal structure of a phosphoribosyltransferase.
|
| |
Biochemistry,
37,
17120-17127.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.J.Focia,
S.P.Craig,
R.Nieves-Alicea,
R.J.Fletterick,
and
A.E.Eakin
(1998).
A 1.4 A crystal structure for the hypoxanthine phosphoribosyltransferase of Trypanosoma cruzi.
|
| |
Biochemistry,
37,
15066-15075.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
V.Sharma,
C.Grubmeyer,
and
J.C.Sacchettini
(1998).
Crystal structure of quinolinic acid phosphoribosyltransferase from Mmycobacterium tuberculosis: a potential TB drug target.
|
| |
Structure,
6,
1587-1599.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Krahn,
J.H.Kim,
M.R.Burns,
R.J.Parry,
H.Zalkin,
and
J.L.Smith
(1997).
Coupled formation of an amidotransferase interdomain ammonia channel and a phosphoribosyltransferase active site.
|
| |
Biochemistry,
36,
11061-11068.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Chen,
D.R.Tomchick,
D.Wolle,
P.Hu,
J.L.Smith,
R.L.Switzer,
and
H.Zalkin
(1997).
Mechanism of the synergistic end-product regulation of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase by nucleotides.
|
| |
Biochemistry,
36,
10718-10726.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

