 |
PDBsum entry 1q7e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.8.3.16
- formyl-CoA transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
formyl-CoA + oxalate = oxalyl-CoA + formate
|
 |
 |
 |
 |
 |
 formyl-CoA
formyl-CoA
|
+
|
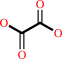 oxalate
oxalate
|
=
|
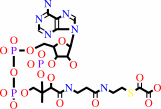 oxalyl-CoA
oxalyl-CoA
|
+
|
 formate
formate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
60:507-511
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of Escherichia coli YfdW, a type III CoA transferase.
|
|
A.Gogos,
J.Gorman,
L.Shapiro.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Crystal structures are reported for free and coenzyme A (CoA) bound forms of the
YfdW protein from Escherichia coli, a representative type III CoA transferase.
The structures reveal a two-domain protomer with interdomain connections forming
a ring-like structure with a large central hole. Two protomers associate to form
a highly intertwined dimer in which the hole of each ring is filled by the
partner molecule. Each protomer binds a single CoA molecule and these
CoA-binding sites are distant from one another in the dimer.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Superposition of four YfdW structures: (i) apo form,
shown in magenta, (ii) bound to CoA, shown in green, (iii) bound
to acetyl-CoA (Gruez et al., 2003[Gruez, A., Roig-Zamboni, V.,
Valencia, C., Campanacci, V. & Cambillau, C. (2003). J. Biol.
Chem. 278, 34582-34586.]), shown in cyan, and (iv) bound to
acetyl-CoA and oxalate (Gruez et al., 2003[Gruez, A.,
Roig-Zamboni, V., Valencia, C., Campanacci, V. & Cambillau, C.
(2003). J. Biol. Chem. 278, 34582-34586.]), shown in gray. CoA
and acetyl-CoA are partly shown at the top of the picture, in
blue and yellow, respectively. Oxalate is depicted in yellow at
the bottom of the picture in an all-atom representation. Note
the shift in conformation of the glycine-rich loop apparently
induced by acetyl-CoA.
|
 |
Figure 4.
Figure 4 Binding of CoA to the putative active site of YfdW. A
CoA omit map contoured at 2  is
shown in magenta. CoA is depicted in blue in an all-atom
representation and the water molecules are shown as red spheres.
All CoA interactions are within the large domain. The adenine
moiety is positioned between the side chains of Arg38, Thr74,
Phe97 and Met105. There are hydrogen-bonding interactions
between adenine N6 and the carbonyl O atom of Leu72 and a
water-mediated hydrogen bond between adenine N7 and the carbonyl
O atom of Ile36. For the ribose moiety, O4' forms a hydrogen
bond to Arg38 NH1 and O2' forms a water-mediated hydrogen bond
to the carbonyl O atom of Ala101. The ribose phosphate moiety
forms hydrogen bonds to His98 N is
shown in magenta. CoA is depicted in blue in an all-atom
representation and the water molecules are shown as red spheres.
All CoA interactions are within the large domain. The adenine
moiety is positioned between the side chains of Arg38, Thr74,
Phe97 and Met105. There are hydrogen-bonding interactions
between adenine N6 and the carbonyl O atom of Leu72 and a
water-mediated hydrogen bond between adenine N7 and the carbonyl
O atom of Ile36. For the ribose moiety, O4' forms a hydrogen
bond to Arg38 NH1 and O2' forms a water-mediated hydrogen bond
to the carbonyl O atom of Ala101. The ribose phosphate moiety
forms hydrogen bonds to His98 N  2
through O9 and O7 and to Lys75 N 2
through O9 and O7 and to Lys75 N  through
O8. The binding of the CoA pantetheine chain is stabilized by
van der Waals interactions with Met200 and several
hydrogen-bonding interactions. AO1 of the pyrophosphate moiety
hydrogen bonds to Arg38 N through
O8. The binding of the CoA pantetheine chain is stabilized by
van der Waals interactions with Met200 and several
hydrogen-bonding interactions. AO1 of the pyrophosphate moiety
hydrogen bonds to Arg38 N  2,
AO4 to the hydroxyl of Tyr139 through two water molecules and
AO5 to Lys137 N 2,
AO4 to the hydroxyl of Tyr139 through two water molecules and
AO5 to Lys137 N 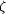 .
The pantetheine hydroxyl group hydrogen bonds to the amide of
His98 and the pantetheine amino PN4 interacts with the carbonyl
O atom of Ala138 and PN8 with the carbonyl O atom of Asn96. The
pantetheine carbonyl O atom PO5 interacts with Asn96 ND2, which
is stabilized by a hydrogen bond to Ser18 OG. The pantetheine
SH group PS1 hydrogen bonds to the amide of Asn17 and is also in
close contact with Asp169 OD2. The SH group also appears to be
stabilized by a helix dipole interaction with the N-terminus of .
The pantetheine hydroxyl group hydrogen bonds to the amide of
His98 and the pantetheine amino PN4 interacts with the carbonyl
O atom of Ala138 and PN8 with the carbonyl O atom of Asn96. The
pantetheine carbonyl O atom PO5 interacts with Asn96 ND2, which
is stabilized by a hydrogen bond to Ser18 OG. The pantetheine
SH group PS1 hydrogen bonds to the amide of Asn17 and is also in
close contact with Asp169 OD2. The SH group also appears to be
stabilized by a helix dipole interaction with the N-terminus of
 -helix
1. -helix
1.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2004,
60,
507-511)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.G.Toyota,
C.L.Berthold,
A.Gruez,
S.Jónsson,
Y.Lindqvist,
C.Cambillau,
and
N.G.Richards
(2008).
Differential substrate specificity and kinetic behavior of Escherichia coli YfdW and Oxalobacter formigenes formyl coenzyme A transferase.
|
| |
J Bacteriol,
190,
2556-2564.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Friedmann,
A.Steindorf,
B.E.Alber,
and
G.Fuchs
(2006).
Properties of succinyl-coenzyme A:L-malate coenzyme A transferase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus.
|
| |
J Bacteriol,
188,
2646-2655.
|
 |
|
|
|
|
 |
S.Friedmann,
B.E.Alber,
and
G.Fuchs
(2006).
Properties of succinyl-coenzyme A:D-citramalate coenzyme A transferase and its role in the autotrophic 3-hydroxypropionate cycle of Chloroflexus aurantiacus.
|
| |
J Bacteriol,
188,
6460-6468.
|
 |
|
|
|
|
 |
K.Savolainen,
P.Bhaumik,
W.Schmitz,
T.J.Kotti,
E.Conzelmann,
R.K.Wierenga,
and
J.K.Hiltunen
(2005).
Alpha-methylacyl-CoA racemase from Mycobacterium tuberculosis. Mutational and structural characterization of the active site and the fold.
|
| |
J Biol Chem,
280,
12611-12620.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Jonsson,
S.Ricagno,
Y.Lindqvist,
and
N.G.Richards
(2004).
Kinetic and mechanistic characterization of the formyl-CoA transferase from Oxalobacter formigenes.
|
| |
J Biol Chem,
279,
36003-36012.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

