 |
PDBsum entry 1pwm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1pwm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of human aldose reductase complexed with NADP and fidarestat
|
|
Structure:
|
 |
Aldose reductase. Chain: a. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli bl21. Expression_system_taxid: 511693.
|
|
Resolution:
|
 |
|
0.92Å
|
R-factor:
|
0.105
|
R-free:
|
0.129
|
|
|
Authors:
|
 |
O.El-Kabbani,C.Darmanin,T.R.Schneider,I.Hazemann,F.Ruiz,M.Oka, A.Joachimiak,C.Schulze-Briese,T.Tomizaki,A.Mitschler,A.Podjarny
|
Key ref:

|
 |
O.El-Kabbani
et al.
(2004).
Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors.
Proteins,
55,
805-813.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
02-Jul-03
|
Release date:
|
24-Feb-04
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P15121
(ALDR_HUMAN) -
Aldo-keto reductase family 1 member B1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
316 a.a.
316 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.21
- aldose reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
an alditol + NAD+ = an aldose + NADH + H+
|
|
2.
|
an alditol + NADP+ = an aldose + NADPH + H+
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 aldose
aldose
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 alditol
alditol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 aldose
aldose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.300
- NADP-retinol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
all-trans-retinol + NADP+ = all-trans-retinal + NADPH + H+
|
 |
 |
 |
 |
 |
all-trans-retinol
Bound ligand (Het Group name = )
matches with 41.38% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
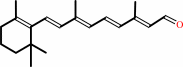 all-trans-retinal
all-trans-retinal
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.1.1.372
- D/L-glyceraldehyde reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
glycerol + NADP+ = L-glyceraldehyde + NADPH + H+
|
|
2.
|
glycerol + NADP+ = D-glyceraldehyde + NADPH + H+
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
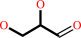 L-glyceraldehyde
L-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 glycerol
glycerol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
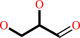 D-glyceraldehyde
D-glyceraldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.1.1.54
- allyl-alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
allyl alcohol + NADP+ = acrolein + NADPH + H+
|
 |
 |
 |
 |
 |
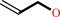 allyl alcohol
allyl alcohol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
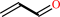 acrolein
acrolein
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
55:805-813
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors.
|
|
O.El-Kabbani,
C.Darmanin,
T.R.Schneider,
I.Hazemann,
F.Ruiz,
M.Oka,
A.Joachimiak,
C.Schulze-Briese,
T.Tomizaki,
A.Mitschler,
A.Podjarny.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The X-ray structures of human aldose reductase holoenzyme in complex with the
inhibitors Fidarestat (SNK-860) and Minalrestat (WAY-509) were determined at
atomic resolutions of 0.92 A and 1.1 A, respectively. The hydantoin and
succinimide moieties of the inhibitors interacted with the conserved
anion-binding site located between the nicotinamide ring of the coenzyme and
active site residues Tyr48, His110, and Trp111. Minalrestat's hydrophobic
isoquinoline ring was bound in an adjacent pocket lined by residues Trp20,
Phe122, and Trp219, with the bromo-fluorobenzyl group inside the
"specificity" pocket. The interactions between Minalrestat's
bromo-fluorobenzyl group and the enzyme include the stacking against the
side-chain of Trp111 as well as hydrogen bonding distances with residues Leu300
and Thr113. The carbamoyl group in Fidarestat formed a hydrogen bond with the
main-chain nitrogen atom of Leu300. The atomic resolution refinement allowed the
positioning of hydrogen atoms and accurate determination of bond lengths of the
inhibitors, coenzyme NADP+ and active-site residue His110. The 1'-position
nitrogen atom in the hydantoin and succinimide moieties of Fidarestat and
Minalrestat, respectively, form a hydrogen bond with the Nepsilon2 atom of His
110. For Fidarestat, the electron density indicated two possible positions for
the H-atom in this bond. Furthermore, both native and anomalous difference maps
indicated the replacement of a water molecule linked to His110 by a Cl-ion.
These observations suggest a mechanism in which Fidarestat is bound protonated
and becomes negatively charged by donating the proton to His110, which may have
important implications on drug design.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 5.
Figure 5. Stereoviews of (a) Fidarestat and (b) Minalrestat
bound into the active site of the human ALR2 holoenzyme.
Residues within 4 Å of the compounds with hydrogen bonds
(yellow solid lines) and close contacts (green solid lines) are
shown.
|
 |
Figure 6.
Figure 6. Proposed mechanism of binding of Fidarestat (SNK-860).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2004,
55,
805-813)
copyright 2004.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.J.Dodson,
and
M.M.Woolfson
(2009).
ACORN2: new developments of the ACORN concept.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
881-891.
|
 |
|
|
|
|
 |
M.Eisenmann,
H.Steuber,
M.Zentgraf,
M.Altenkämper,
R.Ortmann,
J.Perruchon,
G.Klebe,
and
M.Schlitzer
(2009).
Structure-based optimization of aldose reductase inhibitors originating from virtual screening.
|
| |
ChemMedChem,
4,
809-819.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Rakowitz,
G.Piccolruaz,
C.Pirker,
and
B.Matuszczak
(2007).
Novel aldose reductase inhibitors derived from 6-[[(diphenylmethylene)amino]oxy]hexanoic acid.
|
| |
Arch Pharm (Weinheim),
340,
202-208.
|
 |
|
|
|
|
 |
M.Biadene,
I.Hazemann,
A.Cousido,
S.Ginell,
A.Joachimiak,
G.M.Sheldrick,
A.Podjarny,
and
T.R.Schneider
(2007).
The atomic resolution structure of human aldose reductase reveals that rearrangement of a bound ligand allows the opening of the safety-belt loop.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
665-672.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Liu,
B.L.Hanson,
P.Langan,
and
R.E.Viola
(2007).
The effect of deuteration on protein structure: a high-resolution comparison of hydrogenous and perdeuterated haloalkane dehalogenase.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
1000-1008.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.M.Brownlee,
E.Carlson,
A.C.Milne,
E.Pape,
and
D.H.Harrison
(2006).
Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
|
| |
Bioorg Chem,
34,
424-444.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Hazemann,
M.T.Dauvergne,
M.P.Blakeley,
F.Meilleur,
M.Haertlein,
A.Van Dorsselaer,
A.Mitschler,
D.A.Myles,
and
A.Podjarny
(2005).
High-resolution neutron protein crystallography with radically small crystal volumes: application of perdeuteration to human aldose reductase.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1413-1417.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

