 |
PDBsum entry 1ptg

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase (phosphoric diester)
|
PDB id
|
|

|

|
1ptg
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.6.1.13
- phosphatidylinositol diacylglycerol-lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
1-Phosphatidyl-myo-inositol Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol) = 1D-myo-inositol 1,2-cyclic phosphate + a 1,2-diacyl-sn-glycerol
|
 |
 |
 |
 |
 |
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol)
|
=
|
1D-myo-inositol 1,2-cyclic phosphate
Bound ligand (Het Group name = )
matches with 80.00% similarity
|
+
|
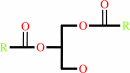 1,2-diacyl-sn-glycerol
1,2-diacyl-sn-glycerol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Embo J
14:3855-3863
(1995)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol.
|
|
D.W.Heinz,
M.Ryan,
T.L.Bullock,
O.H.Griffith.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phosphatidylinositol (PI), once regarded as an obscure component of membranes,
is now recognized as an important reservoir of second messenger precursors and
as an anchor for membrane enzymes. PI-specific phospholipase C (PI-PLC) is the
enzyme that cleaves PI, invoking numerous cellular responses. The crystal
structure of PI-PLC from Bacillus cereus (EC 3.1.4.10) has been solved at 2.6 A
resolution and refined to a crystallographic R factor of 18.7%. The structure
consists of an imperfect (beta alpha)8-barrel similar to that first observed for
triose phosphate isomerase and does not resemble any other known phospholipase
structure. The active site of the enzyme has been identified by determining the
structure of PI-PLC in complex with its inhibitor, myo-inositol, at 2.6 A
resolution (R factor = 19.5%). This substrate-like inhibitor interacts with a
number of residues highly conserved among prokaryotic PI-PLCs. Residues His32
and His82, which are also conserved between prokaryotic and eukaryotic PI-PLCs,
most likely act as general base and acid respectively in a catalytic mechanism
analogous to that observed for ribonucleases.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Jerga,
D.J.Miller,
S.W.White,
and
C.O.Rock
(2009).
Molecular determinants for interfacial binding and conformational change in a soluble diacylglycerol kinase.
|
| |
J Biol Chem,
284,
7246-7254.
|
 |
|
|
|
|
 |
A.Masayama,
K.Tsukada,
C.Ikeda,
H.Nakano,
and
Y.Iwasaki
(2009).
Isolation of phospholipase D mutants having phosphatidylinositol-synthesizing activity with positional specificity on myo-inositol.
|
| |
Chembiochem,
10,
559-564.
|
 |
|
|
|
|
 |
M.Pu,
J.Feng,
A.G.Redfield,
and
M.F.Roberts
(2009).
Enzymology with a spin-labeled phospholipase C: soluble substrate binding by 31P NMR from 0.005 to 11.7 T.
|
| |
Biochemistry,
48,
8282-8284.
|
 |
|
|
|
|
 |
M.Pu,
M.F.Roberts,
and
A.Gershenson
(2009).
Fluorescence correlation spectroscopy of phosphatidylinositol-specific phospholipase C monitors the interplay of substrate and activator lipid binding.
|
| |
Biochemistry,
48,
6835-6845.
|
 |
|
|
|
|
 |
M.Pu,
X.Fang,
A.G.Redfield,
A.Gershenson,
and
M.F.Roberts
(2009).
Correlation of Vesicle Binding and Phospholipid Dynamics with Phospholipase C Activity: INSIGHTS INTO PHOSPHATIDYLCHOLINE ACTIVATION AND SURFACE DILUTION INHIBITION.
|
| |
J Biol Chem,
284,
16099-16107.
|
 |
|
|
|
|
 |
X.Shi,
C.Shao,
X.Zhang,
C.Zambonelli,
A.G.Redfield,
J.F.Head,
B.A.Seaton,
and
M.F.Roberts
(2009).
Modulation of Bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
|
| |
J Biol Chem,
284,
15607-15618.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Shi,
J.F.Liu,
X.M.An,
and
D.C.Liang
(2008).
Crystal structure of glycerophosphodiester phosphodiesterase (GDPD) from Thermoanaerobacter tengcongensis, a metal ion-dependent enzyme: insight into the catalytic mechanism.
|
| |
Proteins,
72,
280-288.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Guo,
X.Zhang,
B.A.Seaton,
and
M.F.Roberts
(2008).
Role of helix B residues in interfacial activation of a bacterial phosphatidylinositol-specific phospholipase C.
|
| |
Biochemistry,
47,
4201-4210.
|
 |
|
|
|
|
 |
C.Shao,
X.Shi,
H.Wehbi,
C.Zambonelli,
J.F.Head,
B.A.Seaton,
and
M.F.Roberts
(2007).
Dimer structure of an interfacially impaired phosphatidylinositol-specific phospholipase C.
|
| |
J Biol Chem,
282,
9228-9235.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Drin,
and
S.Scarlata
(2007).
Stimulation of phospholipase Cbeta by membrane interactions, interdomain movement, and G protein binding--how many ways can you activate an enzyme?
|
| |
Cell Signal,
19,
1383-1392.
|
 |
|
|
|
|
 |
J.T.Stivers,
and
R.Nagarajan
(2006).
Probing enzyme phosphoester interactions by combining mutagenesis and chemical modification of phosphate ester oxygens.
|
| |
Chem Rev,
106,
3443-3467.
|
 |
|
|
|
|
 |
Y.S.Yun,
W.Lee,
S.Shin,
B.H.Oh,
and
K.Y.Choi
(2006).
Arg-158 is critical in both binding the substrate and stabilizing the transition-state oxyanion for the enzymatic reaction of malonamidase E2.
|
| |
J Biol Chem,
281,
40057-40064.
|
 |
|
|
|
|
 |
A.E.Openshaw,
P.R.Race,
H.J.Monzó,
J.A.Vázquez-Boland,
and
M.J.Banfield
(2005).
Crystal structure of SmcL, a bacterial neutral sphingomyelinase C from Listeria.
|
| |
J Biol Chem,
280,
35011-35017.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Z.Wei,
L.A.Zenewicz,
and
H.Goldfine
(2005).
Listeria monocytogenes phosphatidylinositol-specific phospholipase C has evolved for virulence by greatly reduced activity on GPI anchors.
|
| |
Proc Natl Acad Sci U S A,
102,
12927-12931.
|
 |
|
|
|
|
 |
L.Zhao,
H.Liao,
and
M.D.Tsai
(2004).
The catalytic role of aspartate in a short strong hydrogen bond of the Asp274-His32 catalytic dyad in phosphatidylinositol-specific phospholipase C can be substituted by a chloride ion.
|
| |
J Biol Chem,
279,
31995-32000.
|
 |
|
|
|
|
 |
X.Zhang,
H.Wehbi,
and
M.F.Roberts
(2004).
Cross-linking phosphatidylinositol-specific phospholipase C traps two activating phosphatidylcholine molecules on the enzyme.
|
| |
J Biol Chem,
279,
20490-20500.
|
 |
|
|
|
|
 |
G.B.Birrell,
T.O.Zaikova,
A.V.Rukavishnikov,
J.F.Keana,
and
O.H.Griffith
(2003).
Allosteric interactions within subsites of a monomeric enzyme: kinetics of fluorogenic substrates of PI-specific phospholipase C.
|
| |
Biophys J,
84,
3264-3275.
|
 |
|
|
|
|
 |
J.Feng,
W.D.Bradley,
and
M.F.Roberts
(2003).
Optimizing the interfacial binding and activity of a bacterial phosphatidylinositol-specific phospholipase C.
|
| |
J Biol Chem,
278,
24651-24657.
|
 |
|
|
|
|
 |
J.T.Snyder,
A.U.Singer,
M.R.Wing,
T.K.Harden,
and
J.Sondek
(2003).
The pleckstrin homology domain of phospholipase C-beta2 as an effector site for Rac.
|
| |
J Biol Chem,
278,
21099-21104.
|
 |
|
|
|
|
 |
J.Feng,
H.Wehbi,
and
M.F.Roberts
(2002).
Role of tryptophan residues in interfacial binding of phosphatidylinositol-specific phospholipase C.
|
| |
J Biol Chem,
277,
19867-19875.
|
 |
|
|
|
|
 |
A.V.Kravchuk,
L.Zhao,
R.J.Kubiak,
K.S.Bruzik,
and
M.D.Tsai
(2001).
Mechanism of phosphatidylinositol-specific phospholipase C: origin of unusually high nonbridging thio effects.
|
| |
Biochemistry,
40,
5433-5439.
|
 |
|
|
|
|
 |
R.J.Kubiak,
X.Yue,
R.J.Hondal,
C.Mihai,
M.D.Tsai,
and
K.S.Bruzik
(2001).
Involvement of the Arg-Asp-His catalytic triad in enzymatic cleavage of the phosphodiester bond.
|
| |
Biochemistry,
40,
5422-5432.
|
 |
|
|
|
|
 |
I.Leiros,
F.Secundo,
C.Zambonelli,
S.Servi,
and
E.Hough
(2000).
The first crystal structure of a phospholipase D.
|
| |
Structure,
8,
655-667.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.L.Williams
(1999).
Mammalian phosphoinositide-specific phospholipase C.
|
| |
Biochim Biophys Acta,
1441,
255-267.
|
 |
|
|
|
|
 |
T.Bannam,
and
H.Goldfine
(1999).
Mutagenesis of active-site histidines of Listeria monocytogenes phosphatidylinositol-specific phospholipase C: effects on enzyme activity and biological function.
|
| |
Infect Immun,
67,
182-186.
|
 |
|
|
|
|
 |
C.Zhou,
and
M.F.Roberts
(1998).
Nonessential activation and competitive inhibition of bacterial phosphatidylinositol-specific phospholipase C by short-chain phospholipids and analogues.
|
| |
Biochemistry,
37,
16430-16439.
|
 |
|
|
|
|
 |
J.A.Grobler,
and
J.H.Hurley
(1998).
Catalysis by phospholipase C delta1 requires that Ca2+ bind to the catalytic domain, but not the C2 domain.
|
| |
Biochemistry,
37,
5020-5028.
|
 |
|
|
|
|
 |
M.Katan
(1998).
Families of phosphoinositide-specific phospholipase C: structure and function.
|
| |
Biochim Biophys Acta,
1436,
5.
|
 |
|
|
|
|
 |
M.V.Ellis,
S.R.James,
O.Perisic,
C.P.Downes,
R.L.Williams,
and
M.Katan
(1998).
Catalytic domain of phosphoinositide-specific phospholipase C (PLC). Mutational analysis of residues within the active site and hydrophobic ridge of plcdelta1.
|
| |
J Biol Chem,
273,
11650-11659.
|
 |
|
|
|
|
 |
R.J.Hondal,
Z.Zhao,
A.V.Kravchuk,
H.Liao,
S.R.Riddle,
X.Yue,
K.S.Bruzik,
and
M.D.Tsai
(1998).
Mechanism of phosphatidylinositol-specific phospholipase C: a unified view of the mechanism of catalysis.
|
| |
Biochemistry,
37,
4568-4580.
|
 |
|
|
|
|
 |
C.S.Gässler,
M.Ryan,
T.Liu,
O.H.Griffith,
and
D.W.Heinz
(1997).
Probing the roles of active site residues in phosphatidylinositol-specific phospholipase C from Bacillus cereus by site-directed mutagenesis.
|
| |
Biochemistry,
36,
12802-12813.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Zhou,
X.Qian,
and
M.F.Roberts
(1997).
Allosteric activation of phosphatidylinositol-specific phospholipase C: specific phospholipid binding anchors the enzyme to the interface.
|
| |
Biochemistry,
36,
10089-10097.
|
 |
|
|
|
|
 |
C.Zhou,
Y.Wu,
and
M.F.Roberts
(1997).
Activation of phosphatidylinositol-specific phospholipase C toward inositol 1,2-(cyclic)-phosphate.
|
| |
Biochemistry,
36,
347-355.
|
 |
|
|
|
|
 |
D.da Graça Thrige,
J.R.Buur,
and
F.S.Jørgensen
(1997).
Substrate binding and catalytic mechanism in phospholipase C from Bacillus cereus: a molecular mechanics and molecular dynamics study.
|
| |
Biopolymers,
42,
319-336.
|
 |
|
|
|
|
 |
E.Tall,
G.Dormán,
P.Garcia,
L.Runnels,
S.Shah,
J.Chen,
A.Profit,
Q.M.Gu,
A.Chaudhary,
G.D.Prestwich,
and
M.J.Rebecchi
(1997).
Phosphoinositide binding specificity among phospholipase C isozymes as determined by photo-cross-linking to novel substrate and product analogs.
|
| |
Biochemistry,
36,
7239-7248.
|
 |
|
|
|
|
 |
J.H.Hurley,
and
J.A.Grobler
(1997).
Protein kinase C and phospholipase C: bilayer interactions and regulation.
|
| |
Curr Opin Struct Biol,
7,
557-565.
|
 |
|
|
|
|
 |
L.O.Essen,
O.Perisic,
M.Katan,
Y.Wu,
M.F.Roberts,
and
R.L.Williams
(1997).
Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
|
| |
Biochemistry,
36,
1704-1718.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
O.Nosjean,
A.Briolay,
and
B.Roux
(1997).
Mammalian GPI proteins: sorting, membrane residence and functions.
|
| |
Biochim Biophys Acta,
1331,
153-186.
|
 |
|
|
|
|
 |
R.J.Hondal,
S.R.Riddle,
A.V.Kravchuk,
Z.Zhao,
H.Liao,
K.S.Bruzik,
and
M.D.Tsai
(1997).
Phosphatidylinositol-specific phospholipase C: kinetic and stereochemical evidence for an interaction between arginine-69 and the phosphate group of phosphatidylinositol.
|
| |
Biochemistry,
36,
6633-6642.
|
 |
|
|
|
|
 |
T.Liu,
M.Ryan,
F.W.Dahlquist,
and
O.H.Griffith
(1997).
Determination of pKa values of the histidine side chains of phosphatidylinositol-specific phospholipase C from Bacillus cereus by NMR spectroscopy and site-directed mutagenesis.
|
| |
Protein Sci,
6,
1937-1944.
|
 |
|
|
|
|
 |
W.D.Singer,
H.A.Brown,
and
P.C.Sternweis
(1997).
Regulation of eukaryotic phosphatidylinositol-specific phospholipase C and phospholipase D.
|
| |
Annu Rev Biochem,
66,
475-509.
|
 |
|
|
|
|
 |
Y.Wu,
C.Zhou,
and
M.F.Roberts
(1997).
Stereocontrolled syntheses of water-soluble inhibitors of phosphatidylinositol-specific phospholipase C: inhibition enhanced by an interface.
|
| |
Biochemistry,
36,
356-363.
|
 |
|
|
|
|
 |
Y.Wu,
and
M.F.Roberts
(1997).
Phosphatidylinositol-specific phospholipase C cyclic phosphodiesterase activity depends on solvent polarity.
|
| |
Biochemistry,
36,
8514-8521.
|
 |
|
|
|
|
 |
Y.Wu,
O.Perisic,
R.L.Williams,
M.Katan,
and
M.F.Roberts
(1997).
Phosphoinositide-specific phospholipase C delta1 activity toward micellar substrates, inositol 1,2-cyclic phosphate, and other water-soluble substrates: a sequential mechanism and allosteric activation.
|
| |
Biochemistry,
36,
11223-11233.
|
 |
|
|
|
|
 |
D.W.Heinz,
M.Ryan,
M.P.Smith,
L.H.Weaver,
J.F.Keana,
and
O.H.Griffith
(1996).
Crystal structure of phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with glucosaminyl(alpha 1-->6)-D-myo-inositol, an essential fragment of GPI anchors.
|
| |
Biochemistry,
35,
9496-9504.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
G.Venkatakrishnan,
and
J.H.Exton
(1996).
Identification of determinants in the alpha-subunit of Gq required for phospholipase C activation.
|
| |
J Biol Chem,
271,
5066-5072.
|
 |
|
|
|
|
 |
P.Bütikofer,
M.Boschung,
U.Brodbeck,
and
A.K.Menon
(1996).
Phosphatidylinositol hydrolysis by Trypanosoma brucei glycosylphosphatidylinositol phospholipase C.
|
| |
J Biol Chem,
271,
15533-15541.
|
 |
|
|
|
|
 |
R.L.Williams,
and
M.Katan
(1996).
Structural views of phosphoinositide-specific phospholipase C: signalling the way ahead.
|
| |
Structure,
4,
1387-1394.
|
 |
|
|
|
|
 |
S.Scarlata,
R.Gupta,
P.Garcia,
H.Keach,
S.Shah,
C.R.Kasireddy,
R.Bittman,
and
M.J.Rebecchi
(1996).
Inhibition of phospholipase C-delta 1 catalytic activity by sphingomyelin.
|
| |
Biochemistry,
35,
14882-14888.
|
 |
|
|
|
|
 |
M.Hyvönen,
M.J.Macias,
M.Nilges,
H.Oschkinat,
M.Saraste,
and
M.Wilmanns
(1995).
Structure of the binding site for inositol phosphates in a PH domain.
|
| |
EMBO J,
14,
4676-4685.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

