 |
PDBsum entry 1pg0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.10
- methionine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Met) + L-methionine + ATP = L-methionyl-tRNA(Met) + AMP + diphosphate
|
 |
 |
 |
 |
 |
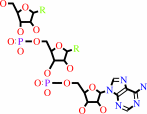 tRNA(Met)
tRNA(Met)
|
+
|
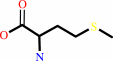 L-methionine
L-methionine
|
+
|
 ATP
ATP
|
=
|
L-methionyl-tRNA(Met)
Bound ligand (Het Group name = )
matches with 76.67% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
332:59-72
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
|
|
T.Crepin,
E.Schmitt,
Y.Mechulam,
P.B.Sampson,
M.D.Vaughan,
J.F.Honek,
S.Blanquet.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Binding of methionine to methionyl-tRNA synthetase (MetRS) is known to promote
conformational changes within the active site. However, the contribution of
these rearrangements to enzyme catalysis is not fully understood. In this study,
several methionine and methionyl adenylate analogues were diffused into crystals
of the monomeric form of Escherichia coli methionyl-tRNA synthetase. The
structures of the corresponding complexes were solved at resolutions below 1.9A
and compared to those of the enzyme free or complexed with methionine. Residues
Y15 and W253 play key roles in the strength of the binding of the amino acid and
of its analogues. Indeed, full motions of these residues are required to recover
the maximum in free energy of binding. Residue Y15 also controls the size of the
hydrophobic pocket where the amino acid side-chain interacts. H301 appears to
participate to the specific recognition of the sulphur atom of methionine.
Complexes with methionyl adenylate analogues illustrate the shielding by MetRS
of the region joining the methionine and adenosine moieties. Finally, the
structure of MetRS complexed to a methionine analogue mimicking the tetrahedral
carbon of the transition state in the aminoacylation reaction was solved. On the
basis of this model, we propose that, in response to the binding of the 3'-end
of tRNA, Y15 moves again in order to deshield the anhydride bond in the natural
adenylate.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Electronic densities associated with Y15 and
W253. A and B show the positions of W253 and Y15 in the free
enzyme[23.] (A) and in the MetRS:Met complex [16.] (B). C-F, The
final 2F[o] -F[c] electron density maps are represented. Each
map is contoured at 1s. C, MetRS:DFM complex; D, MetRS:TFM
complex; E, MetRS:MetI complex; F, MetRS:MetP complex. E, The
two alternative conformations of Y15 in the MetRS:MetI complex
are shown.
|
 |
Figure 4.
Figure 4. Binding of MetSA in the active site of MetRS. A,
Schematic representation of the main electrostatic interactions
between the enzyme and MetSA. Bottom: stereo views of active
site-bound MetSA (B), Metol-AMP (C) and methionine plus
adenosine (D). Only the enzyme residues relevant to the
discussion in the text are drawn.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2003,
332,
59-72)
copyright 2003.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Palencia,
T.Crépin,
M.T.Vu,
T.L.Lincecum,
S.A.Martinis,
and
S.Cusack
(2012).
Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase.
|
| |
Nat Struct Mol Biol,
19,
677-684.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Dalvit,
and
A.Vulpetti
(2011).
Fluorine-protein interactions and ¹⁹F NMR isotropic chemical shifts: An empirical correlation with implications for drug design.
|
| |
ChemMedChem,
6,
104-114.
|
 |
|
|
|
|
 |
H.Ingvarsson,
and
T.Unge
(2010).
Flexibility and communication within the structure of the Mycobacterium smegmatis methionyl-tRNA synthetase.
|
| |
FEBS J,
277,
3947-3962.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.Schmitt,
I.C.Tanrikulu,
T.H.Yoo,
M.Panvert,
D.A.Tirrell,
and
Y.Mechulam
(2009).
Switching from an induced-fit to a lock-and-key mechanism in an aminoacyl-tRNA synthetase with modified specificity.
|
| |
J Mol Biol,
394,
843-851.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
F.Fan,
and
J.S.Blanchard
(2009).
Toward the catalytic mechanism of a cysteine ligase (MshC) from Mycobacterium smegmatis: an enzyme involved in the biosynthetic pathway of mycothiol.
|
| |
Biochemistry,
48,
7150-7159.
|
 |
|
|
|
|
 |
I.C.Tanrikulu,
E.Schmitt,
Y.Mechulam,
W.A.Goddard,
and
D.A.Tirrell
(2009).
Discovery of Escherichia coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo.
|
| |
Proc Natl Acad Sci U S A,
106,
15285-15290.
|
 |
|
|
|
|
 |
L.S.Green,
J.M.Bullard,
W.Ribble,
F.Dean,
D.F.Ayers,
U.A.Ochsner,
N.Janjic,
and
T.C.Jarvis
(2009).
Inhibition of methionyl-tRNA synthetase by REP8839 and effects of resistance mutations on enzyme activity.
|
| |
Antimicrob Agents Chemother,
53,
86-94.
|
 |
|
|
|
|
 |
M.Konno,
T.Sumida,
E.Uchikawa,
Y.Mori,
T.Yanagisawa,
S.Sekine,
and
S.Yokoyama
(2009).
Modeling of tRNA-assisted mechanism of Arg activation based on a structure of Arg-tRNA synthetase, tRNA, and an ATP analog (ANP).
|
| |
FEBS J,
276,
4763-4779.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.W.Tremblay,
F.Fan,
M.W.Vetting,
and
J.S.Blanchard
(2008).
The 1.6 A crystal structure of Mycobacterium smegmatis MshC: the penultimate enzyme in the mycothiol biosynthetic pathway.
|
| |
Biochemistry,
47,
13326-13335.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Ghosh,
and
S.Vishveshwara
(2007).
A study of communication pathways in methionyl- tRNA synthetase by molecular dynamics simulations and structure network analysis.
|
| |
Proc Natl Acad Sci U S A,
104,
15711-15716.
|
 |
|
|
|
|
 |
M.E.Budiman,
M.H.Knaggs,
J.S.Fetrow,
and
R.W.Alexander
(2007).
Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase.
|
| |
Proteins,
68,
670-689.
|
 |
|
|
|
|
 |
A.J.Link,
M.K.Vink,
N.J.Agard,
J.A.Prescher,
C.R.Bertozzi,
and
D.A.Tirrell
(2006).
Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids.
|
| |
Proc Natl Acad Sci U S A,
103,
10180-10185.
|
 |
|
|
|
|
 |
R.Powers,
N.Mirkovic,
S.Goldsmith-Fischman,
T.B.Acton,
Y.Chiang,
Y.J.Huang,
L.Ma,
P.K.Rajan,
J.R.Cort,
M.A.Kennedy,
J.Liu,
B.Rost,
B.Honig,
D.Murray,
and
G.T.Montelione
(2005).
Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes.
|
| |
Protein Sci,
14,
2849-2861.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

