 |
PDBsum entry 1p6c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.8.1
- aryldialkylphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
An aryl dialkyl phosphate + H2O = dialkyl phosphate + an aryl alcohol
|
 |
 |
 |
 |
 |
aryl dialkyl phosphate
Bound ligand (Het Group name = )
matches with 41.18% similarity
|
+
|
H2O
|
=
|
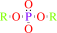 dialkyl phosphate
dialkyl phosphate
|
+
|
aryl alcohol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Am Chem Soc
125:8990-8991
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Enhanced degradation of chemical warfare agents through molecular engineering of the phosphotriesterase active site.
|
|
C.M.Hill,
W.S.Li,
J.B.Thoden,
H.M.Holden,
F.M.Raushel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The bacterial phosphotriesterase has been utilized as a template for the
evolution of improved enzymes for the catalytic decomposition of organophosphate
nerve agents. A combinatorial library of active site mutants was constructed by
randomizing residues His-254, His-257, and Leu-303. The collection of mutant
proteins was screened for the ability to hydrolyze a chromogenic analogue of the
most toxic stereoisomer of the chemical warfare agent, soman. The mutant
H254G/H257W/L303T catalyzed the hydrolysis of the target substrate nearly 3
orders of magnitude faster than the wild-type enzyme. The X-ray crystal
structure was solved in the presence and absence of diisopropyl methyl
phosphonate. The mutant enzyme was ligated to an additional divalent cation at
the active site that was displaced upon the binding of the substrate analogue
inhibitor. These studies demonstrate that substantial changes in substrate
specificity can be achieved by relatively minor changes to the primary amino
acid sequence.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.K.Raynes,
F.G.Pearce,
S.J.Meade,
and
J.A.Gerrard
(2011).
Immobilization of organophosphate hydrolase on an amyloid fibril nanoscaffold: Towards bioremediation and chemical detoxification.
|
| |
Biotechnol Prog,
27,
360-367.
|
 |
|
|
|
|
 |
D.E.Gomes,
R.D.Lins,
P.G.Pascutti,
C.Lei,
and
T.A.Soares
(2010).
The role of nonbonded interactions in the conformational dynamics of organophosphorous hydrolase adsorbed onto functionalized mesoporous silica surfaces.
|
| |
J Phys Chem B,
114,
531-540.
|
 |
|
|
|
|
 |
J.Paramesvaran,
E.G.Hibbert,
A.J.Russell,
and
P.A.Dalby
(2009).
Distributions of enzyme residues yielding mutants with improved substrate specificities from two different directed evolution strategies.
|
| |
Protein Eng Des Sel,
22,
401-411.
|
 |
|
|
|
|
 |
T.Yu,
J.S.Shen,
H.H.Bai,
L.Guo,
J.J.Tang,
Y.B.Jiang,
and
J.W.Xie
(2009).
A photoluminescent nanocrystal-based signaling protocol highly sensitive to nerve agents and highly toxic organophosphate pesticides.
|
| |
Analyst,
134,
2153-2157.
|
 |
|
|
|
|
 |
X.Zhang,
R.Wu,
L.Song,
Y.Lin,
M.Lin,
Z.Cao,
W.Wu,
and
Y.Mo
(2009).
Molecular dynamics simulations of the detoxification of paraoxon catalyzed by phosphotriesterase.
|
| |
J Comput Chem,
30,
2388-2401.
|
 |
|
|
|
|
 |
C.D.Fleming,
C.C.Edwards,
S.D.Kirby,
D.M.Maxwell,
P.M.Potter,
D.M.Cerasoli,
and
M.R.Redinbo
(2007).
Crystal structures of human carboxylesterase 1 in covalent complexes with the chemical warfare agents soman and tabun.
|
| |
Biochemistry,
46,
5063-5071.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.A.Schofield,
C.Westwater,
J.L.Barth,
and
A.A.DiNovo
(2007).
Development of a yeast biosensor-biocatalyst for the detection and biodegradation of the organophosphate paraoxon.
|
| |
Appl Microbiol Biotechnol,
76,
1383-1394.
|
 |
|
|
|
|
 |
J.F.Chaparro-Riggers,
K.M.Polizzi,
and
A.S.Bommarius
(2007).
Better library design: data-driven protein engineering.
|
| |
Biotechnol J,
2,
180-191.
|
 |
|
|
|
|
 |
F.Terrier,
P.Rodriguez-Dafonte,
E.Le Guével,
and
G.Moutiers
(2006).
Revisiting the reactivity of oximate alpha-nucleophiles with electrophilic phosphorus centers. Relevance to detoxification of sarin, soman and DFP under mild conditions.
|
| |
Org Biomol Chem,
4,
4352-4363.
|
 |
|
|
|
|
 |
K.L.Klinkel,
L.A.Kiemele,
D.L.Gin,
and
J.R.Hagadorn
(2006).
Rapid phosphorus triester hydrolysis catalyzed by bimetallic tetrabenzimidazole complexes.
|
| |
Chem Commun (Camb),
(),
2919-2921.
|
 |
|
|
|
|
 |
M.T.Reetz,
J.D.Carballeira,
J.Peyralans,
H.Höbenreich,
A.Maichele,
and
A.Vogel
(2006).
Expanding the substrate scope of enzymes: combining mutations obtained by CASTing.
|
| |
Chemistry,
12,
6031-6038.
|
 |
|
|
|
|
 |
M.T.Reetz,
M.Bocola,
J.D.Carballeira,
D.Zha,
and
A.Vogel
(2005).
Expanding the range of substrate acceptance of enzymes: combinatorial active-site saturation test.
|
| |
Angew Chem Int Ed Engl,
44,
4192-4196.
|
 |
|
|
|
|
 |
R.A.Chica,
N.Doucet,
and
J.N.Pelletier
(2005).
Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design.
|
| |
Curr Opin Biotechnol,
16,
378-384.
|
 |
|
|
|
|
 |
R.Kazlauskas
(2005).
Biological chemistry: enzymes in focus.
|
| |
Nature,
436,
1096-1097.
|
 |
|
|
|
|
 |
C.M.Cho,
A.Mulchandani,
and
W.Chen
(2004).
Altering the substrate specificity of organophosphorus hydrolase for enhanced hydrolysis of chlorpyrifos.
|
| |
Appl Environ Microbiol,
70,
4681-4685.
|
 |
|
|
|
|
 |
T.D.Sutherland,
I.Horne,
K.M.Weir,
C.W.Coppin,
M.R.Williams,
M.Selleck,
R.J.Russell,
and
J.G.Oakeshott
(2004).
Enzymatic bioremediation: from enzyme discovery to applications.
|
| |
Clin Exp Pharmacol Physiol,
31,
817-821.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

