 |
PDBsum entry 1p0c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1p0c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.2
- alcohol dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a primary alcohol + NADP+ = an aldehyde + NADPH + H+
|
 |
 |
 |
 |
 |
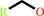 primary alcohol
primary alcohol
|
+
|
 NADP(+)
NADP(+)
|
=
|
 aldehyde
aldehyde
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
330:75-85
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the vertebrate NADP(H)-dependent alcohol dehydrogenase (ADH8).
|
|
A.Rosell,
E.Valencia,
X.Parés,
I.Fita,
J.Farrés,
W.F.Ochoa.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The amphibian enzyme ADH8, previously named class IV-like, is the only known
vertebrate alcohol dehydrogenase (ADH) with specificity towards NADP(H). The
three-dimensional structures of ADH8 and of the binary complex ADH8-NADP(+) have
been now determined and refined to resolutions of 2.2A and 1.8A, respectively.
The coenzyme and substrate specificity of ADH8, that has 50-65% sequence
identity with vertebrate NAD(H)-dependent ADHs, suggest a role in aldehyde
reduction probably as a retinal reductase. The large volume of the
substrate-binding pocket can explain both the high catalytic efficiency of ADH8
with retinoids and the high K(m) value for ethanol. Preference of NADP(H)
appears to be achieved by the presence in ADH8 of the triad Gly223-Thr224-His225
and the recruitment of conserved Lys228, which define a binding pocket for the
terminal phosphate group of the cofactor. NADP(H) binds to ADH8 in an extended
conformation that superimposes well with the NAD(H) molecules found in
NAD(H)-dependent ADH complexes. No additional reshaping of the
dinucleotide-binding site is observed which explains why NAD(H) can also be used
as a cofactor by ADH8. The structural features support the classification of
ADH8 as an independent ADH class.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Stereo views of the cofactor-binding pocket in
the crystal structures of the (a) apo-ADH8 and of the (b)
ADH8-NADP+ complex. Molecular models are represented by solid
sticks with protein atoms colored according to their atom type.
Bound phosphate and glycerol molecules, in the apo-ADH8
structure, and the NADP+ molecule, in the ADH8-NADP+ structure,
are displayed in green. Electron densities, corresponding to the
final 2F[o] -F[c] maps, are also shown, at 1s level, with a
chicken box representation.
|
 |
Figure 4.
Figure 4. LIGPLOT[49.] describing interactions of the NADP+
molecule found in the structure of the ADH8-NADP+ complex. Only
side-chains of residues Thr224, His225 and Lys228 interact with
the terminal phosphate group of the NADP+ cofactor. Thr224 and
His225 are sequence variations specific of ADH8 while Lys228 is
conserved among NAD(H)-dependent ADHs (see in the text).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2003,
330,
75-85)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Ehsani,
M.R.Fernández,
J.A.Biosca,
and
S.Dequin
(2009).
Reversal of coenzyme specificity of 2,3-butanediol dehydrogenase from Saccharomyces cerevisae and in vivo functional analysis.
|
| |
Biotechnol Bioeng,
104,
381-389.
|
 |
|
|
|
|
 |
S.Porté,
E.Valencia,
E.A.Yakovtseva,
E.Borràs,
N.Shafqat,
J.E.Debreczeny,
A.C.Pike,
U.Oppermann,
J.Farrés,
I.Fita,
and
X.Parés
(2009).
Three-dimensional Structure and Enzymatic Function of Proapoptotic Human p53-inducible Quinone Oxidoreductase PIG3.
|
| |
J Biol Chem,
284,
17194-17205.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
R.A.Harris,
J.R.Trudell,
and
S.J.Mihic
(2008).
Ethanol's molecular targets.
|
| |
Sci Signal,
1,
re7.
|
 |
|
|
|
|
 |
M.Domínguez,
R.Alvarez,
E.Borràs,
J.Farrés,
X.Parés,
and
A.R.de Lera
(2006).
Synthesis of enantiopure C3- and C4-hydroxyretinals and their enzymatic reduction by ADH8 from Xenopus laevis.
|
| |
Org Biomol Chem,
4,
155-164.
|
 |
|
|
|
|
 |
C.Montella,
L.Bellsolell,
R.Pérez-Luque,
J.Badía,
L.Baldoma,
M.Coll,
and
J.Aguilar
(2005).
Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli.
|
| |
J Bacteriol,
187,
4957-4966.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Gonzàlez-Duarte,
and
R.Albalat
(2005).
Merging protein, gene and genomic data: the evolution of the MDR-ADH family.
|
| |
Heredity,
95,
184-197.
|
 |
|
|
|
|
 |
S.Watanabe,
T.Kodaki,
and
K.Makino
(2005).
Complete reversal of coenzyme specificity of xylitol dehydrogenase and increase of thermostability by the introduction of structural zinc.
|
| |
J Biol Chem,
280,
10340-10349.
|
 |
|
|
|
|
 |
A.Rosell,
E.Valencia,
W.F.Ochoa,
I.Fita,
X.Parés,
and
J.Farrés
(2003).
Complete reversal of coenzyme specificity by concerted mutation of three consecutive residues in alcohol dehydrogenase.
|
| |
J Biol Chem,
278,
40573-40580.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

