 |
PDBsum entry 1odt

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Cephalosporin c deacetylase mutated, in complex with acetate
|
|
Structure:
|
 |
Cephalosporin c deacetylase. Chain: c, h. Synonym: multi-functional esterase, cah. Engineered: yes. Mutation: yes. Other_details: complex with acetate
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
 Hexamer (from PDB file)
Hexamer (from PDB file)
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.158
|
R-free:
|
0.191
|
|
|
Authors:
|
 |
F.Vincent,S.J.Charnock,K.H.G.Verschueren,J.P.Turkenburg,D.J.Scott, W.A.Offen,S.Roberts,G.Pell,H.J.Gilbert,J.A.Brannigan,G.J.Davies
|
Key ref:

|
 |
F.Vincent
et al.
(2003).
Multifunctional xylooligosaccharide/cephalosporin C deacetylase revealed by the hexameric structure of the Bacillus subtilis enzyme at 1.9A resolution.
J Mol Biol,
330,
593-606.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
20-Feb-03
|
Release date:
|
10-Jul-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P94388
(CAH_BACSU) -
Cephalosporin-C deacetylase from Bacillus subtilis (strain 168)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
318 a.a.
317 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.1.1.41
- cephalosporin-C deacetylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Cephalosporin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
cephalosporin C + H2O = deacetylcephalosporin C + acetate + H+
|
 |
 |
 |
 |
 |
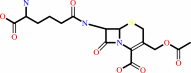 cephalosporin C
cephalosporin C
|
+
|
H2O
|
=
|
deacetylcephalosporin C
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 acetate
acetate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.1.72
- acetylxylan esterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Deacetylation of xylans and xylo-oligosaccharides.
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
330:593-606
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Multifunctional xylooligosaccharide/cephalosporin C deacetylase revealed by the hexameric structure of the Bacillus subtilis enzyme at 1.9A resolution.
|
|
F.Vincent,
S.J.Charnock,
K.H.Verschueren,
J.P.Turkenburg,
D.J.Scott,
W.A.Offen,
S.Roberts,
G.Pell,
H.J.Gilbert,
G.J.Davies,
J.A.Brannigan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Esterases and deacetylases active on carbohydrate ligands have been classified
into 14 families based upon amino acid sequence similarities. Enzymes from
carbohydrate esterase family seven (CE-7) are unusual in that they display
activity towards both acetylated xylooligosaccharides and the antibiotic,
cephalosporin C. The 1.9A structure of the multifunctional CE-7 esterase
(hereinafter CAH) from Bacillus subtilis 168 reveals a classical alpha/beta
hydrolase fold encased within a 32 hexamer. This is the first example of a
hexameric alpha/beta hydrolase and is further evidence of the versatility of
this particular fold, which is used in a wide variety of biological contexts. A
narrow entrance tunnel leads to the centre of the molecule, where the six
active-centre catalytic triads point towards the tunnel interior and thus are
sequestered away from cytoplasmic contents. By analogy to
self-compartmentalising proteases, the tunnel entrance may function to hinder
access of large substrates to the poly-specific active centre. This would
explain the observation that the enzyme is active on a variety of small,
acetylated molecules. The structure of an active site mutant in complex with the
reaction product, acetate, reveals details of the putative oxyanion binding
site, and suggests that substrates bind predominantly through non-specific
contacts with protein hydrophobic residues. Protein residues involved in
catalysis are tethered by interactions with protein excursions from the
canonical alpha/beta hydrolase fold. These excursions also mediate quaternary
structure maintenance, so it would appear that catalytic competence is only
achieved on protein multimerisation. We suggest that the acetyl xylan esterase
(EC 3.1.1.72) and cephalosporin C deacetylase (EC 3.1.1.41) enzymes of the CE-7
family represent a single class of proteins with a multifunctional deacetylase
activity against a range of small substrates.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. The fold of a CAH monomer. (a) Ribbon diagram,
with secondary structure elements from the N to the C terminus
colour ramped blue to red. The active site catalytic triad
residues are depicted as ball-and-stick models, with oxygen
atoms coloured red and nitrogen atoms blue. The extended portion
of the sequence on the top right of the molecule in this
orientation forms the b-sheet-like interface between adjacent
subunits. This and subsequent Figures were drawn with
BOBSCRIPT[72.]/MOLSCRIPT [73.] and raster3D. [74.] (b) Topology
diagram. The a-helices and b-strands are coloured and labelled
as for Figure 3. The b-sheet-like interface-region is shaded in
black. The catalytic triad of Ser181, Asp269 and His298 is
indicated.
|
 |
Figure 8.
Figure 8. Comparison of the oxyanion hole of CAH and
acetyl-xylan esterase AXEII. Divergent stereographic view of the
superposition of the residues involved in the interaction of CAH
with acetate and AXEII[6.] with sulphate. AXEII residues Thr13
and Gln91, which interact with the sulphate ion are in cyan
whilst the equivalent CAH residues (Tyr91 and Gln182)
interacting with the acetate are in white. This Figure was drawn
using PyMOL [76.] (http://www.pymol.org).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2003,
330,
593-606)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Widmann,
P.B.Juhl,
and
J.Pleiss
(2010).
Structural classification by the Lipase Engineering Database: a case study of Candida antarctica lipase A.
|
| |
BMC Genomics,
11,
123.
|
 |
|
|
|
|
 |
E.J.Taylor,
T.M.Gloster,
J.P.Turkenburg,
F.Vincent,
A.M.Brzozowski,
C.Dupont,
F.Shareck,
M.S.Centeno,
J.A.Prates,
V.Puchart,
L.M.Ferreira,
C.M.Fontes,
P.Biely,
and
G.J.Davies
(2006).
Structure and activity of two metal ion-dependent acetylxylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases.
|
| |
J Biol Chem,
281,
10968-10975.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.A.van den Broek,
R.M.Lloyd,
G.Beldman,
J.C.Verdoes,
B.V.McCleary,
and
A.G.Voragen
(2005).
Cloning and characterization of arabinoxylan arabinofuranohydrolase-D3 (AXHd3) from Bifidobacterium adolescentis DSM20083.
|
| |
Appl Microbiol Biotechnol,
67,
641-647.
|
 |
|
|
|
|
 |
F.Vincent,
D.Yates,
E.Garman,
G.J.Davies,
and
J.A.Brannigan
(2004).
The three-dimensional structure of the N-acetylglucosamine-6-phosphate deacetylase, NagA, from Bacillus subtilis: a member of the urease superfamily.
|
| |
J Biol Chem,
279,
2809-2816.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

