 |
PDBsum entry 1odb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Metal binding protein
|
PDB id
|
|

|

|
1odb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Metal binding protein
|
 |
|
Title:
|
 |
The crystal structure of human s100a12 - copper complex
|
|
Structure:
|
 |
Calgranulin c. Chain: a, b, c, d, e, f. Synonym: s100a12, cagc, p6, cgrp, neutrophil s100 protein, calcium- binding protein in amniotic fluid 1, caaf1. Engineered: yes. Other_details: ca2+ and cu2+ bound form
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Tissue: blood. Cell: granulocyte. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
 Dimer (from PDB file)
Dimer (from PDB file)
|
|
Resolution:
|
 |
|
2.19Å
|
R-factor:
|
0.188
|
R-free:
|
0.226
|
|
|
Authors:
|
 |
O.V.Moroz,A.A.Antson,S.J.Grist,N.J.Maitland,G.G.Dodson,K.S.Wilson, E.M.Lukanidin,I.B.Bronstein
|
Key ref:

|
 |
O.V.Moroz
et al.
(2003).
Structure of the human S100A12-copper complex: implications for host-parasite defence.
Acta Crystallogr D Biol Crystallogr,
59,
859-867.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
15-Feb-03
|
Release date:
|
12-Jun-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P80511
(S10AC_HUMAN) -
Protein S100-A12 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
92 a.a.
91 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
59:859-867
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of the human S100A12-copper complex: implications for host-parasite defence.
|
|
O.V.Moroz,
A.A.Antson,
S.J.Grist,
N.J.Maitland,
G.G.Dodson,
K.S.Wilson,
E.Lukanidin,
I.B.Bronstein.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
S100A12 is a member of the S100 family of EF-hand calcium-modulated proteins.
Together with S100A8 and S100A9, it belongs to the calgranulin subfamily, i.e.
it is mainly expressed in granulocytes, although there is an increasing body of
evidence of expression in keratinocytes and psoriatic lesions. As well as being
linked to inflammation, allergy and neuritogenesis, S100A12 is involved in
host-parasite response, as are the other two calgranulins. Recent data suggest
that the function of the S100-family proteins is modulated not only by calcium,
but also by other metals such as zinc and copper. Previously, the structure of
human S100A12 in low-calcium and high-calcium structural forms, crystallized in
space groups R3 and P2(1), respectively, has been reported. Here, the structure
of S100A12 in complex with copper (space group P2(1)2(1)2; unit-cell parameters
a = 70.6, b = 119.0, c = 90.2 A) refined at 2.19 A resolution is reported.
Comparison of anomalous difference electron-density maps calculated with data
collected with radiation of wavelengths 1.37 and 1.65 A shows that each monomer
binds a single copper ion. The copper binds at an equivalent site to that at
which another S100 protein, S100A7, binds zinc. The results suggest that copper
binding may be essential for the functional role of S100A12 and probably the
other calgranulins in the early immune response.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 (a)  ( (
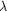 )
plots for copper (in blue) and for calcium (in red). Dotted
lines correspond to the wavelengths for set I ( )
plots for copper (in blue) and for calcium (in red). Dotted
lines correspond to the wavelengths for set I ( 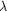 [I]
= 1.37 Å) and for data set II ( [I]
= 1.37 Å) and for data set II ( 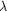 [II]
= 1.65 Å). (b) Anomalous difference electron-density maps for
data sets I ( [II]
= 1.65 Å). (b) Anomalous difference electron-density maps for
data sets I ( 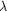 =
1.37 Å) and II ( =
1.37 Å) and II ( 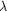 =
1.65 Å). Electron density for data set I is contoured at the 10 =
1.65 Å). Electron density for data set I is contoured at the 10
 level
and is shown in red. Electron density for data set II is
contoured at the 5 level
and is shown in red. Electron density for data set II is
contoured at the 5  level
and is shown in green. This figure and Fig. 4-were generated
using QUANTA (Molecular Simulations, Inc.). level
and is shown in green. This figure and Fig. 4-were generated
using QUANTA (Molecular Simulations, Inc.).
|
 |
Figure 6.
Figure 6 Superposition of the copper-binding site of S100A12
(PDB code [287]1odb ), the zinc-binding site of S100A7
([288]2psr ) and corresponding regions of S100A8 ([289]1mr8 ),
S100A9 ([290]1irj ) and S100B ([291]1mho ) shown in
ball-and-stick representation. S100A12 is shown in blue, S100A8
in green, S100A9 in orange, S100A7 in yellow and S100B in
violet. Copper ion from the S100A12-copper complex structure is
in blue, zinc ion from the S100A7-zinc complex is in yellow. The
figure was generated using MOLSCRIPT (Kraulis, 1991[292]
[Kraulis, P. (1991). J. Appl. Cryst. 24,
946-950.]-[293][bluearr.gif] ) and Raster3D (Merritt & Murphy,
1994[294] [Merritt, E. A. & Murphy, M. E. P. (1994). Acta Cryst.
D50, 869-873.]-[295][bluearr.gif] ).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2003,
59,
859-867)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Unno,
T.Kawasaki,
H.Takahara,
C.W.Heizmann,
and
K.Kizawa
(2011).
Refined crystal structures of human Ca(2+)/Zn(2+)-binding S100A3 protein characterized by two disulfide bridges.
|
| |
J Mol Biol,
408,
477-490.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Ostendorp,
J.Diez,
C.W.Heizmann,
and
G.Fritz
(2011).
The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
|
| |
Biochim Biophys Acta,
1813,
1083-1091.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Grabarek
(2011).
Insights into modulation of calcium signaling by magnesium in calmodulin, troponin C and related EF-hand proteins.
|
| |
Biochim Biophys Acta,
1813,
913-921.
|
 |
|
|
|
|
 |
G.Fritz,
H.M.Botelho,
L.A.Morozova-Roche,
and
C.M.Gomes
(2010).
Natural and amyloid self-assembly of S100 proteins: structural basis of functional diversity.
|
| |
FEBS J,
277,
4578-4590.
|
 |
|
|
|
|
 |
T.E.Kehl-Fie,
and
E.P.Skaar
(2010).
Nutritional immunity beyond iron: a role for manganese and zinc.
|
| |
Curr Opin Chem Biol,
14,
218-224.
|
 |
|
|
|
|
 |
J.Pietzsch,
and
S.Hoppmann
(2009).
Human S100A12: a novel key player in inflammation?
|
| |
Amino Acids,
36,
381-389.
|
 |
|
|
|
|
 |
K.Hsu,
C.Champaiboon,
B.D.Guenther,
B.S.Sorenson,
A.Khammanivong,
K.F.Ross,
C.L.Geczy,
and
M.C.Herzberg
(2009).
ANTI-INFECTIVE PROTECTIVE PROPERTIES OF S100 CALGRANULINS.
|
| |
Antiinflamm Antiallergy Agents Med Chem,
8,
290-305.
|
 |
|
|
|
|
 |
O.V.Moroz,
W.Burkitt,
H.Wittkowski,
W.He,
A.Ianoul,
V.Novitskaya,
J.Xie,
O.Polyakova,
I.K.Lednev,
A.Shekhtman,
P.J.Derrick,
P.Bjoerk,
D.Foell,
and
I.B.Bronstein
(2009).
Both Ca2+ and Zn2+ are essential for S100A12 protein -oligomerization and function.
|
| |
BMC Biochem,
10,
11.
|
 |
|
|
|
|
 |
T.H.Charpentier,
P.T.Wilder,
M.A.Liriano,
K.M.Varney,
E.Pozharski,
A.D.MacKerell,
A.Coop,
E.A.Toth,
and
D.J.Weber
(2008).
Divalent metal ion complexes of S100B in the absence and presence of pentamidine.
|
| |
J Mol Biol,
382,
56-73.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Larsen,
I.B.Bronstein,
O.Dahl,
T.Wentzel-Larsen,
E.K.Kristoffersen,
and
M.K.Fagerhol
(2007).
Quantification of S100A12 (EN-RAGE) in blood varies with sampling method, calcium and heparin.
|
| |
Scand J Immunol,
65,
192-201.
|
 |
|
|
|
|
 |
A.J.Fielding,
S.Fox,
G.L.Millhauser,
M.Chattopadhyay,
P.M.Kroneck,
G.Fritz,
G.R.Eaton,
and
S.S.Eaton
(2006).
Electron spin relaxation of copper(II) complexes in glassy solution between 10 and 120 K.
|
| |
J Magn Reson,
179,
92.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

