 |
PDBsum entry 1obc

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.4
- leucine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Leu) + L-leucine + ATP = L-leucyl-tRNA(Leu) + AMP + diphosphate
|
 |
 |
 |
 |
 |
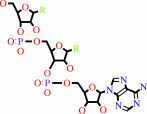 tRNA(Leu)
tRNA(Leu)
|
+
|
L-leucine
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
 ATP
ATP
|
=
|
L-leucyl-tRNA(Leu)
Bound ligand (Het Group name = )
matches with 70.37% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Mol Cell
11:951-963
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and mechanistic basis of pre- and posttransfer editing by leucyl-tRNA synthetase.
|
|
T.L.Lincecum,
M.Tukalo,
A.Yaremchuk,
R.S.Mursinna,
A.M.Williams,
B.S.Sproat,
W.Van Den Eynde,
A.Link,
S.Van Calenbergh,
M.Grøtli,
S.A.Martinis,
S.Cusack.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The aminoacyl-tRNA synthetases link tRNAs with their cognate amino acid. In some
cases, their fidelity relies on hydrolytic editing that destroys incorrectly
activated amino acids or mischarged tRNAs. We present structures of leucyl-tRNA
synthetase complexed with analogs of the distinct pre- and posttransfer editing
substrates. The editing active site binds the two different substrates using a
single amino acid discriminatory pocket while preserving the same mode of
adenine recognition. This suggests a similar mechanism of hydrolysis for both
editing substrates that depends on a key, completely conserved aspartic acid,
which interacts with the alpha-amino group of the noncognate amino acid and
positions both substrates for hydrolysis. Our results demonstrate the economy by
which a single active site accommodates two distinct substrates in a
proofreading process critical to the fidelity of protein synthesis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Editing Reactions, Editing Substrates, and
Sequence Conservation in the Editing Domain(A) LeuRS
aminoacylation and editing reactions. Editing reactions are
indicated by the dashed arrows. Although tRNA has been shown to
be a cofactor for pretransfer editing by IleRS (Baldwin and
Berg, 1966), its role in LeuRS editing is unknown.(B) Diagrams
of the analogs used in this work of LeuRS pre- and posttransfer
editing substrates for the case of noncognate norvaline (Nva).
Left: posttransfer substrate analog,
2′-(L-norvalyl)amino-2′-deoxyadenosine (Nva2AA), mimicking
the 3′ end of the aminoacyl-2′-ester Nva-tRNA^Leu. Right:
pretransfer substrate analog,
5′-O-[N-(L-norvalyl)sulphamoyl]adenosine (NvaAMS), a sulfamoyl
analog of norvalyl-adenylate. In each case, the labile ester
linkages were replaced by a nonhydrolyzable amino linkage to
permit structural studies.(C) Alignment of conserved regions
within the editing (CP1) domain of selected LeuRS (L), ValRS
(V), and IleRS (I) enzymes. The “threonine-rich region”
contains two highly conserved threonines (arrowed) discussed in
the text. In the second region, separated by a bracket, a
conserved glycine-rich loop is followed by a completely
conserved aspartic acid (arrowed) that was mutated to alanine.
Abbreviations: Sc, S. cerevisiae; Ce, Caenorhabditis elegans;
Hs, Homo sapiens; Nc, Neurospora crassa; Ec, E. coli; Tt,
Thermus thermophilus; Bs, Bacillus subtilis; Gs, Geobacillus
stearothermophilus; Sa, Staphylococcus aureus; cyt, cytoplasmic;
mit, mitochondrial.
|
 |
Figure 2.
Figure 2. Electron Density of the Editing Substrates(A)
Simulated omit map (Brunger et al., 1998) for the pretransfer
substrate analog (NvaAMS) in the editing (top) and synthetic
(bottom) active site. Resolution is 2.2 Å. In both
molecules, the ribose is in the C2′ endo conformation.(B)
Location of the NvaAMS in the synthetic and editing active sites
of LeuRSTT.(C) Unbiased difference map (2.0 Å resolution)
for the posttransfer editing substrate analog (Nva2AA) in the
editing site. The ribose is in the C3′ endo conformation.(D)
Competitive inhibition of E. coli LeuRS editing of Ile-tRNA^Leu
by Nva2AA. Editing of Ile-tRNA^Leu by wild-type E. coli LeuRS in
the absence of inhibitor exhibited a K[M] of 0.2 μM.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Mol Cell
(2003,
11,
951-963)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Palencia,
T.Crépin,
M.T.Vu,
T.L.Lincecum,
S.A.Martinis,
and
S.Cusack
(2012).
Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase.
|
| |
Nat Struct Mol Biol,
19,
677-684.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Li,
L.Yu,
and
Q.Huang
(2011).
Molecular trigger for pre-transfer editing pathway in Valyl-tRNA synthetase: A molecular dynamics simulation study.
|
| |
J Mol Model,
17,
555-564.
|
 |
|
|
|
|
 |
X.Chen,
J.J.Ma,
M.Tan,
P.Yao,
Q.H.Hu,
G.Eriani,
and
E.D.Wang
(2011).
Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase.
|
| |
Nucleic Acids Res,
39,
235-247.
|
 |
|
|
|
|
 |
D.Moras
(2010).
Proofreading in translation: dynamics of the double-sieve model.
|
| |
Proc Natl Acad Sci U S A,
107,
21949-21950.
|
 |
|
|
|
|
 |
J.Ling,
and
D.Söll
(2010).
Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site.
|
| |
Proc Natl Acad Sci U S A,
107,
4028-4033.
|
 |
|
|
|
|
 |
M.A.Wouters,
S.W.Fan,
and
N.L.Haworth
(2010).
Disulfides as redox switches: from molecular mechanisms to functional significance.
|
| |
Antioxid Redox Signal,
12,
53-91.
|
 |
|
|
|
|
 |
M.Tan,
B.Zhu,
X.L.Zhou,
R.He,
X.Chen,
G.Eriani,
and
E.D.Wang
(2010).
tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase.
|
| |
J Biol Chem,
285,
3235-3244.
|
 |
|
|
|
|
 |
S.A.Martinis,
and
M.T.Boniecki
(2010).
The balance between pre- and post-transfer editing in tRNA synthetases.
|
| |
FEBS Lett,
584,
455-459.
|
 |
|
|
|
|
 |
T.Hussain,
V.Kamarthapu,
S.P.Kruparani,
M.V.Deshmukh,
and
R.Sankaranarayanan
(2010).
Mechanistic insights into cognate substrate discrimination during proofreading in translation.
|
| |
Proc Natl Acad Sci U S A,
107,
22117-22121.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.L.Zhou,
M.Tan,
M.Wang,
X.Chen,
and
E.D.Wang
(2010).
Post-transfer editing by a eukaryotic leucyl-tRNA synthetase resistant to the broad-spectrum drug AN2690.
|
| |
Biochem J,
430,
325-333.
|
 |
|
|
|
|
 |
A.P.Mascarenhas,
and
S.A.Martinis
(2009).
A glycine hinge for tRNA-dependent translocation of editing substrates to prevent errors by leucyl-tRNA synthetase.
|
| |
FEBS Lett,
583,
3443-3447.
|
 |
|
|
|
|
 |
B.Zhu,
P.Yao,
M.Tan,
G.Eriani,
and
E.D.Wang
(2009).
tRNA-independent Pretransfer Editing by Class I Leucyl-tRNA Synthetase.
|
| |
J Biol Chem,
284,
3418-3424.
|
 |
|
|
|
|
 |
D.Wen,
M.M.Vecchi,
S.Gu,
L.Su,
J.Dolnikova,
Y.M.Huang,
S.F.Foley,
E.Garber,
N.Pederson,
and
W.Meier
(2009).
Discovery and investigation of misincorporation of serine at asparagine positions in recombinant proteins expressed in Chinese hamster ovary cells.
|
| |
J Biol Chem,
284,
32686-32694.
|
 |
|
|
|
|
 |
J.Ling,
B.R.So,
S.S.Yadavalli,
H.Roy,
S.Shoji,
K.Fredrick,
K.Musier-Forsyth,
and
M.Ibba
(2009).
Resampling and editing of mischarged tRNA prior to translation elongation.
|
| |
Mol Cell,
33,
654-660.
|
 |
|
|
|
|
 |
J.Ling,
N.Reynolds,
and
M.Ibba
(2009).
Aminoacyl-tRNA synthesis and translational quality control.
|
| |
Annu Rev Microbiol,
63,
61-78.
|
 |
|
|
|
|
 |
R.A.Hellmann,
and
S.A.Martinis
(2009).
Defects in Transient tRNA Translocation Bypass tRNA Synthetase Quality Control Mechanisms.
|
| |
J Biol Chem,
284,
11478-11484.
|
 |
|
|
|
|
 |
S.F.Ataide,
T.E.Rogers,
and
M.Ibba
(2009).
The CCA anticodon specifies separate functions inside and outside translation in Bacillus cereus.
|
| |
RNA Biol,
6,
479-487.
|
 |
|
|
|
|
 |
S.W.Fan,
R.A.George,
N.L.Haworth,
L.L.Feng,
J.Y.Liu,
and
M.A.Wouters
(2009).
Conformational changes in redox pairs of protein structures.
|
| |
Protein Sci,
18,
1745-1765.
|
 |
|
|
|
|
 |
Y.L.Pang,
and
S.A.Martinis
(2009).
A paradigm shift for the amino acid editing mechanism of human cytoplasmic leucyl-tRNA synthetase.
|
| |
Biochemistry,
48,
8958-8964.
|
 |
|
|
|
|
 |
B.Ruan,
S.Palioura,
J.Sabina,
L.Marvin-Guy,
S.Kochhar,
R.A.Larossa,
and
D.Söll
(2008).
Quality control despite mistranslation caused by an ambiguous genetic code.
|
| |
Proc Natl Acad Sci U S A,
105,
16502-16507.
|
 |
|
|
|
|
 |
C.S.Francklyn
(2008).
DNA polymerases and aminoacyl-tRNA synthetases: shared mechanisms for ensuring the fidelity of gene expression.
|
| |
Biochemistry,
47,
11695-11703.
|
 |
|
|
|
|
 |
J.L.Hsu,
and
S.A.Martinis
(2008).
A Flexible peptide tether controls accessibility of a unique C-terminal RNA-binding domain in leucyl-tRNA synthetases.
|
| |
J Mol Biol,
376,
482-491.
|
 |
|
|
|
|
 |
K.E.Splan,
K.Musier-Forsyth,
M.T.Boniecki,
and
S.A.Martinis
(2008).
In vitro assays for the determination of aminoacyl-tRNA synthetase editing activity.
|
| |
Methods,
44,
119-128.
|
 |
|
|
|
|
 |
M.T.Boniecki,
M.T.Vu,
A.K.Betha,
and
S.A.Martinis
(2008).
CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase.
|
| |
Proc Natl Acad Sci U S A,
105,
19223-19228.
|
 |
|
|
|
|
 |
N.G.Richards
(2008).
Shining a light on post-translational modification.
|
| |
HFSP J,
2,
57-60.
|
 |
|
|
|
|
 |
P.Yao,
B.Zhu,
S.Jaeger,
G.Eriani,
and
E.D.Wang
(2008).
Recognition of tRNALeu by Aquifex aeolicus leucyl-tRNA synthetase during the aminoacylation and editing steps.
|
| |
Nucleic Acids Res,
36,
2728-2738.
|
 |
|
|
|
|
 |
P.Yao,
X.L.Zhou,
R.He,
M.Q.Xue,
Y.G.Zheng,
Y.F.Wang,
and
E.D.Wang
(2008).
Unique residues crucial for optimal editing in yeast cytoplasmic Leucyl-tRNA synthetase are revealed by using a novel knockout yeast strain.
|
| |
J Biol Chem,
283,
22591-22600.
|
 |
|
|
|
|
 |
S.S.Yadavalli,
K.Musier-Forsyth,
and
M.Ibba
(2008).
The return of pretransfer editing in protein synthesis.
|
| |
Proc Natl Acad Sci U S A,
105,
19031-19032.
|
 |
|
|
|
|
 |
A.K.Betha,
A.M.Williams,
and
S.A.Martinis
(2007).
Isolated CP1 domain of Escherichia coli leucyl-tRNA synthetase is dependent on flanking hinge motifs for amino acid editing activity.
|
| |
Biochemistry,
46,
6258-6267.
|
 |
|
|
|
|
 |
E.A.Lemke,
D.Summerer,
B.H.Geierstanger,
S.M.Brittain,
and
P.G.Schultz
(2007).
Control of protein phosphorylation with a genetically encoded photocaged amino acid.
|
| |
Nat Chem Biol,
3,
769-772.
|
 |
|
|
|
|
 |
F.L.Rock,
W.Mao,
A.Yaremchuk,
M.Tukalo,
T.Crépin,
H.Zhou,
Y.K.Zhang,
V.Hernandez,
T.Akama,
S.J.Baker,
J.J.Plattner,
L.Shapiro,
S.A.Martinis,
S.J.Benkovic,
S.Cusack,
and
M.R.Alley
(2007).
An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site.
|
| |
Science,
316,
1759-1761.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Ling,
H.Roy,
and
M.Ibba
(2007).
Mechanism of tRNA-dependent editing in translational quality control.
|
| |
Proc Natl Acad Sci U S A,
104,
72-77.
|
 |
|
|
|
|
 |
J.Ling,
S.S.Yadavalli,
and
M.Ibba
(2007).
Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine.
|
| |
RNA,
13,
1881-1886.
|
 |
|
|
|
|
 |
M.C.Hartman,
K.Josephson,
C.W.Lin,
and
J.W.Szostak
(2007).
An expanded set of amino Acid analogs for the ribosomal translation of unnatural peptides.
|
| |
PLoS ONE,
2,
e972.
|
 |
|
|
|
|
 |
M.T.Vu,
and
S.A.Martinis
(2007).
A unique insert of leucyl-tRNA synthetase is required for aminoacylation and not amino acid editing.
|
| |
Biochemistry,
46,
5170-5176.
|
 |
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2007).
Structure of the AlaX-M trans-editing enzyme from Pyrococcus horikoshii.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
390-400.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.W.Lue,
and
S.O.Kelley
(2007).
A single residue in leucyl-tRNA synthetase affecting amino acid specificity and tRNA aminoacylation.
|
| |
Biochemistry,
46,
4466-4472.
|
 |
|
|
|
|
 |
A.M.Williams,
and
S.A.Martinis
(2006).
Mutational unmasking of a tRNA-dependent pathway for preventing genetic code ambiguity.
|
| |
Proc Natl Acad Sci U S A,
103,
3586-3591.
|
 |
|
|
|
|
 |
C.I.Jones,
A.C.Spencer,
J.L.Hsu,
L.L.Spremulli,
S.A.Martinis,
M.DeRider,
and
P.F.Agris
(2006).
A counterintuitive Mg2+-dependent and modification-assisted functional folding of mitochondrial tRNAs.
|
| |
J Mol Biol,
362,
771-786.
|
 |
|
|
|
|
 |
D.Summerer,
S.Chen,
N.Wu,
A.Deiters,
J.W.Chin,
and
P.G.Schultz
(2006).
A genetically encoded fluorescent amino acid.
|
| |
Proc Natl Acad Sci U S A,
103,
9785-9789.
|
 |
|
|
|
|
 |
H.M.Sasaki,
S.Sekine,
T.Sengoku,
R.Fukunaga,
M.Hattori,
Y.Utsunomiya,
C.Kuroishi,
S.Kuramitsu,
M.Shirouzu,
and
S.Yokoyama
(2006).
Structural and mutational studies of the amino acid-editing domain from archaeal/eukaryal phenylalanyl-tRNA synthetase.
|
| |
Proc Natl Acad Sci U S A,
103,
14744-14749.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.L.Hsu,
S.B.Rho,
K.M.Vannella,
and
S.A.Martinis
(2006).
Functional divergence of a unique C-terminal domain of leucyl-tRNA synthetase to accommodate its splicing and aminoacylation roles.
|
| |
J Biol Chem,
281,
23075-23082.
|
 |
|
|
|
|
 |
J.S.Weinger,
and
S.A.Strobel
(2006).
Participation of the tRNA A76 hydroxyl groups throughout translation.
|
| |
Biochemistry,
45,
5939-5948.
|
 |
|
|
|
|
 |
S.Hati,
B.Ziervogel,
J.Sternjohn,
F.C.Wong,
M.C.Nagan,
A.E.Rosen,
P.G.Siliciano,
J.W.Chihade,
and
K.Musier-Forsyth
(2006).
Pre-transfer editing by class II prolyl-tRNA synthetase: role of aminoacylation active site in "selective release" of noncognate amino acids.
|
| |
J Biol Chem,
281,
27862-27872.
|
 |
|
|
|
|
 |
T.Crepin,
A.Yaremchuk,
M.Tukalo,
and
S.Cusack
(2006).
Structures of two bacterial prolyl-tRNA synthetases with and without a cis-editing domain.
|
| |
Structure,
14,
1511-1525.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.Hussain,
S.P.Kruparani,
B.Pal,
A.C.Dock-Bregeon,
S.Dwivedi,
M.R.Shekar,
K.Sureshbabu,
and
R.Sankaranarayanan
(2006).
Post-transfer editing mechanism of a D-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea.
|
| |
EMBO J,
25,
4152-4162.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
V.A.Karkhanis,
M.T.Boniecki,
K.Poruri,
and
S.A.Martinis
(2006).
A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria.
|
| |
J Biol Chem,
281,
33217-33225.
|
 |
|
|
|
|
 |
H.Roy,
J.Ling,
J.Alfonzo,
and
M.Ibba
(2005).
Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase.
|
| |
J Biol Chem,
280,
38186-38192.
|
 |
|
|
|
|
 |
I.Gruic-Sovulj,
N.Uter,
T.Bullock,
and
J.J.Perona
(2005).
tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting class I aminoacyl-tRNA synthetase.
|
| |
J Biol Chem,
280,
23978-23986.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Tukalo,
A.Yaremchuk,
R.Fukunaga,
S.Yokoyama,
and
S.Cusack
(2005).
The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation.
|
| |
Nat Struct Mol Biol,
12,
923-930.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.W.Zhao,
B.Zhu,
R.Hao,
M.G.Xu,
G.Eriani,
and
E.D.Wang
(2005).
Leucyl-tRNA synthetase from the ancestral bacterium Aquifex aeolicus contains relics of synthetase evolution.
|
| |
EMBO J,
24,
1430-1439.
|
 |
|
|
|
|
 |
R.Fukunaga,
R.Ishitani,
O.Nureki,
and
S.Yokoyama
(2005).
Crystallization of leucyl-tRNA synthetase complexed with tRNALeu from the archaeon Pyrococcus horikoshii.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
61,
30-32.
|
 |
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2005).
Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition.
|
| |
Nat Struct Mol Biol,
12,
915-922.
|
 |
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2005).
Structural basis for non-cognate amino acid discrimination by the valyl-tRNA synthetase editing domain.
|
| |
J Biol Chem,
280,
29937-29945.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Korencic,
I.Ahel,
J.Schelert,
M.Sacher,
B.Ruan,
C.Stathopoulos,
P.Blum,
M.Ibba,
and
D.Söll
(2004).
A freestanding proofreading domain is required for protein synthesis quality control in Archaea.
|
| |
Proc Natl Acad Sci U S A,
101,
10260-10265.
|
 |
|
|
|
|
 |
H.Roy,
J.Ling,
M.Irnov,
and
M.Ibba
(2004).
Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase.
|
| |
EMBO J,
23,
4639-4648.
|
 |
|
|
|
|
 |
K.D.Tardif,
and
J.Horowitz
(2004).
Functional group recognition at the aminoacylation and editing sites of E. coli valyl-tRNA synthetase.
|
| |
RNA,
10,
493-503.
|
 |
|
|
|
|
 |
M.G.Xu,
M.W.Zhao,
and
E.D.Wang
(2004).
Leucyl-tRNA synthetase from the hyperthermophilic bacterium Aquifex aeolicus recognizes minihelices.
|
| |
J Biol Chem,
279,
32151-32158.
|
 |
|
|
|
|
 |
R.Fukunaga,
S.Fukai,
R.Ishitani,
O.Nureki,
and
S.Yokoyama
(2004).
Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L-valine.
|
| |
J Biol Chem,
279,
8396-8402.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2004).
Crystallization and preliminary X-ray crystallographic study of the editing domain of Thermus thermophilus isoleucyl-tRNA synthetase complexed with pre- and post-transfer editing-substrate analogues.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1900-1902.
|
 |
|
|
|
|
 |
R.Fukunaga,
and
S.Yokoyama
(2004).
Crystallization and preliminary X-ray crystallographic study of leucyl-tRNA synthetase from the archaeon Pyrococcus horikoshii.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1916-1918.
|
 |
|
|
|
|
 |
W.Kim,
A.George,
M.Evans,
and
V.P.Conticello
(2004).
Cotranslational incorporation of a structurally diverse series of proline analogues in an Escherichia coli expression system.
|
| |
Chembiochem,
5,
928-936.
|
 |
|
|
|
|
 |
Y.G.Zheng,
H.Wei,
C.Ling,
F.Martin,
G.Eriani,
and
E.D.Wang
(2004).
Two distinct domains of the beta subunit of Aquifex aeolicus leucyl-tRNA synthetase are involved in tRNA binding as revealed by a three-hybrid selection.
|
| |
Nucleic Acids Res,
32,
3294-3303.
|
 |
|
|
|
|
 |
B.E.Nordin,
and
P.Schimmel
(2003).
Transiently misacylated tRNA is a primer for editing of misactivated adenylates by class I aminoacyl-tRNA synthetases.
|
| |
Biochemistry,
42,
12989-12997.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

