
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Naphthalene 1,2-dioxygenase with oxidized rieske iron sulphur center site.
|
|
Structure:
|
 |
Naphthalene 1,2-dioxygenase alpha subunit. Chain: a. Synonym: naphthalene 1,2-dioxygenase isp alpha, ndob, nahac. Engineered: yes. Naphthalene 1,2-dioxygenase beta subunit. Chain: b. Synonym: naphthalene 1,2-dioxygenase isp beta, ndoc, nahad. Engineered: yes
|
|
Source:
|
 |
Pseudomonas putida. Organism_taxid: 303. Strain: ncib 9816-4. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Hexamer (from PDB file)
Hexamer (from PDB file)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.208
|
R-free:
|
0.242
|
|
|
Authors:
|
 |
A.Karlsson,J.V.Parales,R.E.Parales,D.T.Gibson,H.Eklund,S.Ramaswamy
|
Key ref:

|
 |
A.Karlsson
et al.
(2003).
Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron.
Science,
299,
1039-1042.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
05-Nov-02
|
Release date:
|
20-Feb-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain A:
E.C.1.14.12.12
- naphthalene 1,2-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Aromatic 1,2-Dioxygenases
|
 |
 |
 |
 |
 |
Reaction:
|
 |
naphthalene + NADH + O2 + H+ = (1R,2S)-1,2-dihydronaphthalene-1,2-diol + NAD+
|
 |
 |
 |
 |
 |
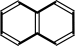 naphthalene
naphthalene
|
+
|
 NADH
NADH
|
+
|
 O2
O2
|
+
|
H(+)
|
=
|
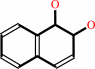 (1R,2S)-1,2-dihydronaphthalene-1,2-diol
(1R,2S)-1,2-dihydronaphthalene-1,2-diol
|
+
|
 NAD(+)
NAD(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
299:1039-1042
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of naphthalene dioxygenase: side-on binding of dioxygen to iron.
|
|
A.Karlsson,
J.V.Parales,
R.E.Parales,
D.T.Gibson,
H.Eklund,
S.Ramaswamy.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Binding of oxygen to iron is exploited in several biological and chemical
processes. Although computational and spectroscopic results have suggested
side-on binding, only end-on binding of oxygen to iron has been observed in
crystal structures. We have determined structures of naphthalene dioxygenase
that show a molecular oxygen species bound to the mononuclear iron in a side-on
fashion. In a complex with substrate and dioxygen, the dioxygen molecule is
lined up for an attack on the double bond of the aromatic substrate. The
structures reported here provide the basis for a reaction mechanism and for the
high stereospecificity of the reaction catalyzed by naphthalene dioxygenase.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. (All panels are stereopairs.) (A) Binding of
naphthalene at the active site of NDO. The gray 2F[obs] -
F[calc] map is contoured at 1.0  . (B)
Binding of dioxygen to the mononuclear iron in the absence of
substrate. The gray 2F[obs] - F[calc] map is contoured at 1.15 . (B)
Binding of dioxygen to the mononuclear iron in the absence of
substrate. The gray 2F[obs] - F[calc] map is contoured at 1.15
 and the
green F[obs] - F[calc] map (computed before the dioxygen
molecule was modeled) at 3.8 × RMS (root mean square). (C)
Binding of oxygen to the mononuclear iron in the presence of
indole. The gray 2F[obs] - F[calc] map is contoured at 1.0 and the
green F[obs] - F[calc] map (computed before the dioxygen
molecule was modeled) at 3.8 × RMS (root mean square). (C)
Binding of oxygen to the mononuclear iron in the presence of
indole. The gray 2F[obs] - F[calc] map is contoured at 1.0  and the
green F[obs] - F[calc] map (computed before the dioxygen
molecule was modeled) at 4.0 × RMS. (D) Naphthalene
cis-dihydrodiol bound to the active site of NDO. The gray
2F[obs] - F[calc] map is contoured at 1.0 and the
green F[obs] - F[calc] map (computed before the dioxygen
molecule was modeled) at 4.0 × RMS. (D) Naphthalene
cis-dihydrodiol bound to the active site of NDO. The gray
2F[obs] - F[calc] map is contoured at 1.0  .
Superposition of the product complex and the substrate complex
shows that the positions of the rings in the product and the
substrate are similar. The product cannot move any closer to the
iron, as it would bring the O from the product into van der
Waals short contact with the His ligand of the Fe. The current
distance between the O and the N of His is 2.9 Å. Color
code: yellow, carbon; blue, nitrogen; red, oxygen; purple, iron. .
Superposition of the product complex and the substrate complex
shows that the positions of the rings in the product and the
substrate are similar. The product cannot move any closer to the
iron, as it would bring the O from the product into van der
Waals short contact with the His ligand of the Fe. The current
distance between the O and the N of His is 2.9 Å. Color
code: yellow, carbon; blue, nitrogen; red, oxygen; purple, iron.
|
 |
Figure 2.
Fig. 2. (A) Scheme showing how the different structures in Fig.
1 can be arranged to follow the dihydroxylation reaction
catalyzed by NDO. Naphthalene and indole are both substrates for
the enzyme, and we have used them interchangeably in different
studies of the enzyme (23). For simplicity we show only
naphthalene here. The structures are as follows: 1, the resting
enzyme with oxidized Rieske center and ferrous active site
[Protein Data Bank (PDB) code 1O7H]; 2, the reduced enzyme (PDB
code 1O7W); 3, binary dioxgen complex (PDB code 1O7M); 4, binary
substrate complex, structures with both indole and naphthalene
[PDB codes 1EG9 (indole) and 1O7G (naphthalene)]; 5, ternary
substrate dioxygen species, structure with indole (PDB code
1O7N); and 6, product naphthalene cis-1,2-dihydrodiol (PDB code
1O7P). [2Fe-2S] refers to the nearest Rieske iron-sulfur
cluster. (B) Chemical steps in the dioxygenation reaction
carried out by Rieske dioxygenases.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(2003,
299,
1039-1042)
copyright 2003.
|
|
| |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|

|
| |
Figure 2: It is has been shown spectroscopically by the John Lipscomb group, during the catalytic cycle, the substrate binds before dioxygen to the active site of the enzyme.
S. Ramaswamy.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Cho,
S.Jeon,
S.A.Wilson,
L.V.Liu,
E.A.Kang,
J.J.Braymer,
M.H.Lim,
B.Hedman,
K.O.Hodgson,
J.S.Valentine,
E.I.Solomon,
and
W.Nam
(2011).
Structure and reactivity of a mononuclear non-haem iron(III)-peroxo complex.
|
| |
Nature,
478,
502-505.
|
 |
|
|
|
|
 |
J.Seo,
S.I.Kang,
D.Won,
M.Kim,
J.Y.Ryu,
S.W.Kang,
B.H.Um,
C.H.Pan,
J.H.Ahn,
Y.Chong,
R.A.Kanaly,
J.Han,
and
H.G.Hur
(2011).
Absolute configuration-dependent epoxide formation from isoflavan-4-ol stereoisomers by biphenyl dioxygenase of Pseudomonas pseudoalcaligenes strain KF707.
|
| |
Appl Microbiol Biotechnol,
89,
1773-1782.
|
 |
|
|
|
|
 |
A.Mukherjee,
M.A.Cranswick,
M.Chakrabarti,
T.K.Paine,
K.Fujisawa,
E.Münck,
and
L.Que
(2010).
Oxygen activation at mononuclear nonheme iron centers: a superoxo perspective.
|
| |
Inorg Chem,
49,
3618-3628.
|
 |
|
|
|
|
 |
C.Cavazza,
C.Bochot,
P.Rousselot-Pailley,
P.Carpentier,
M.V.Cherrier,
L.Martin,
C.Marchi-Delapierre,
J.C.Fontecilla-Camps,
and
S.Ménage
(2010).
Crystallographic snapshots of the reaction of aromatic C-H with O(2) catalysed by a protein-bound iron complex.
|
| |
Nat Chem,
2,
1069-1076.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Bilis,
and
M.Louloudi
(2010).
The Catalytic Function of Nonheme Iron (III) Complex for Hydrocarbon Oxidation.
|
| |
Bioinorg Chem Appl,
(),
0.
|
 |
|
|
|
|
 |
J.Seo,
S.I.Kang,
J.Y.Ryu,
Y.J.Lee,
K.D.Park,
M.Kim,
D.Won,
H.Y.Park,
J.H.Ahn,
Y.Chong,
R.A.Kanaly,
J.Han,
and
H.G.Hur
(2010).
Location of flavone B-ring controls regioselectivity and stereoselectivity of naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4.
|
| |
Appl Microbiol Biotechnol,
86,
1451-1462.
|
 |
|
|
|
|
 |
M.Martinho,
G.Blain,
and
F.Banse
(2010).
Activation of dioxygen by a mononuclear non-heme iron complex: characterization of a Fe(III)(OOH) intermediate.
|
| |
Dalton Trans,
39,
1630-1634.
|
 |
|
|
|
|
 |
M.Morikawa
(2010).
Dioxygen activation responsible for oxidation of aliphatic and aromatic hydrocarbon compounds: current state and variants.
|
| |
Appl Microbiol Biotechnol,
87,
1595-1603.
|
 |
|
|
|
|
 |
O.Kweon,
S.J.Kim,
J.P.Freeman,
J.Song,
S.Baek,
and
C.E.Cerniglia
(2010).
Substrate Specificity and Structural Characteristics of the Novel Rieske Nonheme Iron Aromatic Ring-Hydroxylating Oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1.
|
| |
MBio,
1,
0.
|
 |
|
|
|
|
 |
P.L.Holland
(2010).
Metal-dioxygen and metal-dinitrogen complexes: where are the electrons?
|
| |
Dalton Trans,
39,
5415-5425.
|
 |
|
|
|
|
 |
S.Iwai,
B.Chai,
W.J.Sul,
J.R.Cole,
S.A.Hashsham,
and
J.M.Tiedje
(2010).
Gene-targeted-metagenomics reveals extensive diversity of aromatic dioxygenase genes in the environment.
|
| |
ISME J,
4,
279-285.
|
 |
|
|
|
|
 |
A.M.Orville,
G.T.Lountos,
S.Finnegan,
G.Gadda,
and
R.Prabhakar
(2009).
Crystallographic, spectroscopic, and computational analysis of a flavin C4a-oxygen adduct in choline oxidase.
|
| |
Biochemistry,
48,
720-728.
|
 |
|
|
|
|
 |
E.I.Solomon,
S.D.Wong,
L.V.Liu,
A.Decker,
and
M.S.Chow
(2009).
Peroxo and oxo intermediates in mononuclear nonheme iron enzymes and related active sites.
|
| |
Curr Opin Chem Biol,
13,
99.
|
 |
|
|
|
|
 |
F.Musat,
A.Galushko,
J.Jacob,
F.Widdel,
M.Kube,
R.Reinhardt,
H.Wilkes,
B.Schink,
and
R.Rabus
(2009).
Anaerobic degradation of naphthalene and 2-methylnaphthalene by strains of marine sulfate-reducing bacteria.
|
| |
Environ Microbiol,
11,
209-219.
|
 |
|
|
|
|
 |
J.K.Capyk,
I.D'Angelo,
N.C.Strynadka,
and
L.D.Eltis
(2009).
Characterization of 3-ketosteroid 9{alpha}-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis.
|
| |
J Biol Chem,
284,
9937-9946.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.F.Pacios,
V.M.Campos,
I.Merino,
and
L.Gómez
(2009).
Structures and thermodynamics of biphenyl dihydrodiol stereoisomers and their metabolites in the enzymatic degradation of arene xenobiotics.
|
| |
J Comput Chem,
30,
2420-2432.
|
 |
|
|
|
|
 |
M.Tarasev,
S.Pullela,
and
D.P.Ballou
(2009).
Distal end of 105-125 loop--a putative reductase binding domain of phthalate dioxygenase.
|
| |
Arch Biochem Biophys,
487,
10-18.
|
 |
|
|
|
|
 |
Y.Feng,
C.Y.Ke,
G.Xue,
and
L.Que
(2009).
Bio-inspired arene cis-dihydroxylation by a non-haem iron catalyst modeling the action of naphthalene dioxygenase.
|
| |
Chem Commun (Camb),
(),
50-52.
|
 |
|
|
|
|
 |
C.R.Simmons,
K.Krishnamoorthy,
S.L.Granett,
D.J.Schuller,
J.E.Dominy,
T.P.Begley,
M.H.Stipanuk,
and
P.A.Karplus
(2008).
A putative Fe2+-bound persulfenate intermediate in cysteine dioxygenase.
|
| |
Biochemistry,
47,
11390-11392.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
E.G.Kovaleva,
and
J.D.Lipscomb
(2008).
Versatility of biological non-heme Fe(II) centers in oxygen activation reactions.
|
| |
Nat Chem Biol,
4,
186-193.
|
 |
|
|
|
|
 |
J.D.Lipscomb
(2008).
Mechanism of extradiol aromatic ring-cleaving dioxygenases.
|
| |
Curr Opin Struct Biol,
18,
644-649.
|
 |
|
|
|
|
 |
L.Que,
and
W.B.Tolman
(2008).
Biologically inspired oxidation catalysis.
|
| |
Nature,
455,
333-340.
|
 |
|
|
|
|
 |
O.Kagami,
K.Shindo,
A.Kyojima,
K.Takeda,
H.Ikenaga,
K.Furukawa,
and
N.Misawa
(2008).
Protein engineering on biphenyl dioxygenase for conferring activity to convert 7-hydroxyflavone and 5,7-dihydroxyflavone (chrysin).
|
| |
J Biosci Bioeng,
106,
121-127.
|
 |
|
|
|
|
 |
P.C.Bruijnincx,
G.van Koten,
and
R.J.Klein Gebbink
(2008).
Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: recent developments in enzymology and modeling studies.
|
| |
Chem Soc Rev,
37,
2716-2744.
|
 |
|
|
|
|
 |
S.Boxhammer,
S.Glaser,
A.Kühl,
A.K.Wagner,
and
C.L.Schmidt
(2008).
Characterization of the recombinant Rieske [2Fe-2S] proteins HcaC and YeaW from E. coli.
|
| |
Biometals,
21,
459-467.
|
 |
|
|
|
|
 |
T.D.Bugg,
and
S.Ramaswamy
(2008).
Non-heme iron-dependent dioxygenases: unravelling catalytic mechanisms for complex enzymatic oxidations.
|
| |
Curr Opin Chem Biol,
12,
134-140.
|
 |
|
|
|
|
 |
T.Ohta,
S.Chakrabarty,
J.D.Lipscomb,
and
E.I.Solomon
(2008).
Near-IR MCD of the nonheme ferrous active site in naphthalene 1,2-dioxygenase: correlation to crystallography and structural insight into the mechanism of Rieske dioxygenases.
|
| |
J Am Chem Soc,
130,
1601-1610.
|
 |
|
|
|
|
 |
C.A.Joseph,
and
M.J.Maroney
(2007).
Cysteine dioxygenase: structure and mechanism.
|
| |
Chem Commun (Camb),
(),
3338-3349.
|
 |
|
|
|
|
 |
D.J.Ferraro,
E.N.Brown,
C.L.Yu,
R.E.Parales,
D.T.Gibson,
and
S.Ramaswamy
(2007).
Structural investigations of the ferredoxin and terminal oxygenase components of the biphenyl 2,3-dioxygenase from Sphingobium yanoikuyae B1.
|
| |
BMC Struct Biol,
7,
10.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.G.Kovaleva,
and
J.D.Lipscomb
(2007).
Crystal structures of Fe2+ dioxygenase superoxo, alkylperoxo, and bound product intermediates.
|
| |
Science,
316,
453-457.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
E.G.Kovaleva,
M.B.Neibergall,
S.Chakrabarty,
and
J.D.Lipscomb
(2007).
Finding intermediates in the O2 activation pathways of non-heme iron oxygenases.
|
| |
Acc Chem Res,
40,
475-483.
|
 |
|
|
|
|
 |
E.L.Ang,
J.P.Obbard,
and
H.Zhao
(2007).
Probing the molecular determinants of aniline dioxygenase substrate specificity by saturation mutagenesis.
|
| |
FEBS J,
274,
928-939.
|
 |
|
|
|
|
 |
G.Anilkumar,
B.Bitterlich,
F.G.Gelalcha,
M.K.Tse,
and
M.Beller
(2007).
An efficient biomimetic Fe-catalyzed epoxidation of olefins using hydrogen peroxide.
|
| |
Chem Commun (Camb),
(),
289-291.
|
 |
|
|
|
|
 |
J.Jakoncic,
Y.Jouanneau,
C.Meyer,
and
V.Stojanoff
(2007).
The crystal structure of the ring-hydroxylating dioxygenase from Sphingomonas CHY-1.
|
| |
FEBS J,
274,
2470-2481.
|
 |
|
|
|
|
 |
K.Shindo,
Y.Shindo,
T.Hasegawa,
A.Osawa,
O.Kagami,
K.Furukawa,
and
N.Misawa
(2007).
Synthesis of highly hydroxylated aromatics by evolved biphenyl dioxygenase and subsequent dihydrodiol dehydrogenase.
|
| |
Appl Microbiol Biotechnol,
75,
1063-1069.
|
 |
|
|
|
|
 |
L.Gómez-Gil,
P.Kumar,
D.Barriault,
J.T.Bolin,
M.Sylvestre,
and
L.D.Eltis
(2007).
Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B-356 as a potent polychlorinated biphenyl-degrading enzyme.
|
| |
J Bacteriol,
189,
5705-5715.
|
 |
|
|
|
|
 |
M.A.Carrondo,
I.Bento,
P.M.Matias,
and
P.F.Lindley
(2007).
Crystallographic evidence for dioxygen interactions with iron proteins.
|
| |
J Biol Inorg Chem,
12,
429-442.
|
 |
|
|
|
|
 |
M.B.Neibergall,
A.Stubna,
Y.Mekmouche,
E.Münck,
and
J.D.Lipscomb
(2007).
Hydrogen peroxide dependent cis-dihydroxylation of benzoate by fully oxidized benzoate 1,2-dioxygenase.
|
| |
Biochemistry,
46,
8004-8016.
|
 |
|
|
|
|
 |
M.P.Jensen,
A.M.Payeras,
A.T.Fiedler,
M.Costas,
J.Kaizer,
A.Stubna,
E.Münck,
and
L.Que
(2007).
Kinetic analysis of the conversion of nonheme (alkylperoxo)iron(III) species to iron(IV) complexes.
|
| |
Inorg Chem,
46,
2398-2408.
|
 |
|
|
|
|
 |
M.S.Seo,
J.Y.Kim,
J.Annaraj,
Y.Kim,
Y.M.Lee,
S.J.Kim,
J.Kim,
and
W.Nam
(2007).
[Mn(tmc)(O2)]+: a side-on peroxido manganese(III) complex bearing a non-heme ligand.
|
| |
Angew Chem Int Ed Engl,
46,
377-380.
|
 |
|
|
|
|
 |
P.F.Widboom,
E.N.Fielding,
Y.Liu,
and
S.D.Bruner
(2007).
Structural basis for cofactor-independent dioxygenation in vancomycin biosynthesis.
|
| |
Nature,
447,
342-345.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Chakrabarty,
R.N.Austin,
D.Deng,
J.T.Groves,
and
J.D.Lipscomb
(2007).
Radical intermediates in monooxygenase reactions of rieske dioxygenases.
|
| |
J Am Chem Soc,
129,
3514-3515.
|
 |
|
|
|
|
 |
D.J.Ferraro,
A.L.Okerlund,
J.C.Mowers,
and
S.Ramaswamy
(2006).
Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
|
| |
J Bacteriol,
188,
6986-6994.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.R.Boyd,
and
T.D.Bugg
(2006).
Arene cis-dihydrodiol formation: from biology to application.
|
| |
Org Biomol Chem,
4,
181-192.
|
 |
|
|
|
|
 |
H.Sugimoto,
S.Oda,
T.Otsuki,
T.Hino,
T.Yoshida,
and
Y.Shiro
(2006).
Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
|
| |
Proc Natl Acad Sci U S A,
103,
2611-2616.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Bento,
M.A.Carrondo,
and
P.F.Lindley
(2006).
Reduction of dioxygen by enzymes containing copper.
|
| |
J Biol Inorg Chem,
11,
539-547.
|
 |
|
|
|
|
 |
K.Lee
(2006).
p-hydroxylation reactions catalyzed by naphthalene dioxygenase.
|
| |
FEMS Microbiol Lett,
255,
316-320.
|
 |
|
|
|
|
 |
M.R.Bukowski,
P.Comba,
A.Lienke,
C.Limberg,
C.Lopez de Laorden,
R.Mas-Ballesté,
M.Merz,
and
L.Que
(2006).
Catalytic epoxidation and 1,2-dihydroxylation of olefins with bispidine-iron(II)/H2O2 systems.
|
| |
Angew Chem Int Ed Engl,
45,
3446-3449.
|
 |
|
|
|
|
 |
M.Tarasev,
A.Pinto,
D.Kim,
S.J.Elliott,
and
D.P.Ballou
(2006).
The "bridging" aspartate 178 in phthalate dioxygenase facilitates interactions between the Rieske center and the iron(II)--mononuclear center.
|
| |
Biochemistry,
45,
10208-10216.
|
 |
|
|
|
|
 |
P.D.Oldenburg,
C.Y.Ke,
A.A.Tipton,
A.A.Shteinman,
and
L.Que
(2006).
A structural and functional model for dioxygenases with a 2-His-1-carboxylate triad.
|
| |
Angew Chem Int Ed Engl,
45,
7975-7978.
|
 |
|
|
|
|
 |
R.Mas-Ballesté,
M.Costas,
T.van den Berg,
and
L.Que
(2006).
Ligand topology effects on olefin oxidations by bio-inspired [FeII(N2Py2)] catalysts.
|
| |
Chemistry,
12,
7489-7500.
|
 |
|
|
|
|
 |
S.Eswaramoorthy,
J.B.Bonanno,
S.K.Burley,
and
S.Swaminathan
(2006).
Mechanism of action of a flavin-containing monooxygenase.
|
| |
Proc Natl Acad Sci U S A,
103,
9832-9837.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Karlsson,
J.V.Parales,
R.E.Parales,
D.T.Gibson,
H.Eklund,
and
S.Ramaswamy
(2005).
NO binding to naphthalene dioxygenase.
|
| |
J Biol Inorg Chem,
10,
483-489.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.G.Keenan,
T.Leungsakul,
B.F.Smets,
M.A.Mori,
D.E.Henderson,
and
T.K.Wood
(2005).
Protein engineering of the archetypal nitroarene dioxygenase of Ralstonia sp. strain U2 for activity on aminonitrotoluenes and dinitrotoluenes through alpha-subunit residues leucine 225, phenylalanine 350, and glycine 407.
|
| |
J Bacteriol,
187,
3302-3310.
|
 |
|
|
|
|
 |
B.M.Martins,
T.Svetlitchnaia,
and
H.Dobbek
(2005).
2-Oxoquinoline 8-monooxygenase oxygenase component: active site modulation by Rieske-[2Fe-2S] center oxidation/reduction.
|
| |
Structure,
13,
817-824.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Bagnéris,
R.Cammack,
and
J.R.Mason
(2005).
Subtle difference between benzene and toluene dioxygenases of Pseudomonas putida.
|
| |
Appl Environ Microbiol,
71,
1570-1580.
|
 |
|
|
|
|
 |
D.Bourgeois,
and
A.Royant
(2005).
Advances in kinetic protein crystallography.
|
| |
Curr Opin Struct Biol,
15,
538-547.
|
 |
|
|
|
|
 |
E.V.Kudrik,
A.Theodoridis,
R.van Eldik,
and
S.V.Makarov
(2005).
Kinetics and mechanism of the Co(II)-assisted oxidation of thioureas by dioxygen.
|
| |
Dalton Trans,
(),
1117-1122.
|
 |
|
|
|
|
 |
J.Annaraj,
Y.Suh,
M.S.Seo,
S.O.Kim,
and
W.Nam
(2005).
Mononuclear nonheme ferric-peroxo complex in aldehyde deformylation.
|
| |
Chem Commun (Camb),
(),
4529-4531.
|
 |
|
|
|
|
 |
J.D.Lipscomb,
and
B.M.Hoffman
(2005).
Allosteric control of O2 reactivity in Rieske oxygenases.
|
| |
Structure,
13,
684-685.
|
 |
|
|
|
|
 |
J.Han,
S.Y.Kim,
J.Jung,
Y.Lim,
J.H.Ahn,
S.I.Kim,
and
H.G.Hur
(2005).
Epoxide formation on the aromatic B ring of flavanone by biphenyl dioxygenase of Pseudomonas pseudoalcaligenes KF707.
|
| |
Appl Environ Microbiol,
71,
5354-5361.
|
 |
|
|
|
|
 |
K.D.Koehntop,
J.P.Emerson,
and
L.Que
(2005).
The 2-His-1-carboxylate facial triad: a versatile platform for dioxygen activation by mononuclear non-heme iron(II) enzymes.
|
| |
J Biol Inorg Chem,
10,
87-93.
|
 |
|
|
|
|
 |
L.Gakhar,
Z.A.Malik,
C.C.Allen,
D.A.Lipscomb,
M.J.Larkin,
and
S.Ramaswamy
(2005).
Structure and increased thermostability of Rhodococcus sp. naphthalene 1,2-dioxygenase.
|
| |
J Bacteriol,
187,
7222-7231.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.L.Neidig,
and
E.I.Solomon
(2005).
Structure-function correlations in oxygen activating non-heme iron enzymes.
|
| |
Chem Commun (Camb),
(),
5843-5863.
|
 |
|
|
|
|
 |
R.E.Parales,
R.Huang,
C.L.Yu,
J.V.Parales,
F.K.Lee,
D.J.Lessner,
M.M.Ivkovic-Jensen,
W.Liu,
R.Friemann,
S.Ramaswamy,
and
D.T.Gibson
(2005).
Purification, characterization, and crystallization of the components of the nitrobenzene and 2-nitrotoluene dioxygenase enzyme systems.
|
| |
Appl Environ Microbiol,
71,
3806-3814.
|
 |
|
|
|
|
 |
T.Leungsakul,
B.G.Keenan,
H.Yin,
B.F.Smets,
and
T.K.Wood
(2005).
Saturation mutagenesis of 2,4-DNT dioxygenase of Burkholderia sp. strain DNT for enhanced dinitrotoluene degradation.
|
| |
Biotechnol Bioeng,
92,
416-426.
|
 |
|
|
|
|
 |
X.Dong,
S.Fushinobu,
E.Fukuda,
T.Terada,
S.Nakamura,
K.Shimizu,
H.Nojiri,
T.Omori,
H.Shoun,
and
T.Wakagi
(2005).
Crystal structure of the terminal oxygenase component of cumene dioxygenase from Pseudomonas fluorescens IP01.
|
| |
J Bacteriol,
187,
2483-2490.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.G.Keenan,
T.Leungsakul,
B.F.Smets,
and
T.K.Wood
(2004).
Saturation mutagenesis of Burkholderia cepacia R34 2,4-dinitrotoluene dioxygenase at DntAc valine 350 for synthesizing nitrohydroquinone, methylhydroquinone, and methoxyhydroquinone.
|
| |
Appl Environ Microbiol,
70,
3222-3231.
|
 |
|
|
|
|
 |
L.P.Wackett
(2004).
Stable isotope probing in biodegradation research.
|
| |
Trends Biotechnol,
22,
153-154.
|
 |
|
|
|
|
 |
P.Hlavica
(2004).
Models and mechanisms of O-O bond activation by cytochrome P450. A critical assessment of the potential role of multiple active intermediates in oxidative catalysis.
|
| |
Eur J Biochem,
271,
4335-4360.
|
 |
|
|
|
|
 |
M.Zielinski,
S.Kahl,
H.J.Hecht,
and
B.Hofer
(2003).
Pinpointing biphenyl dioxygenase residues that are crucial for substrate interaction.
|
| |
J Bacteriol,
185,
6976-6980.
|
 |
|
|
|
|
 |
Z.M.Beharry,
D.M.Eby,
E.D.Coulter,
R.Viswanathan,
E.L.Neidle,
R.S.Phillips,
and
D.M.Kurtz
(2003).
Histidine ligand protonation and redox potential in the rieske dioxygenases: role of a conserved aspartate in anthranilate 1,2-dioxygenase.
|
| |
Biochemistry,
42,
13625-13636.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

