 |
PDBsum entry 1o2d

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1o2d
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.1
- alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a primary alcohol + NAD+ = an aldehyde + NADH + H+
|
|
2.
|
a secondary alcohol + NAD+ = a ketone + NADH + H+
|
|
 |
 |
 |
 |
 |
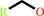 primary alcohol
primary alcohol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
 aldehyde
aldehyde
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
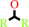 secondary alcohol
secondary alcohol
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
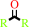 ketone
ketone
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+) or Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
54:174-177
(2004)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of an iron-containing 1,3-propanediol dehydrogenase (TM0920) from Thermotoga maritima at 1.3 A resolution.
|
|
R.Schwarzenbacher,
F.von Delft,
J.M.Canaves,
L.S.Brinen,
X.Dai,
A.M.Deacon,
M.A.Elsliger,
S.Eshaghi,
R.Floyd,
A.Godzik,
C.Grittini,
S.K.Grzechnik,
C.Guda,
L.Jaroszewski,
C.Karlak,
H.E.Klock,
E.Koesema,
J.S.Kovarik,
A.Kreusch,
P.Kuhn,
S.A.Lesley,
D.McMullan,
T.M.McPhillips,
M.A.Miller,
M.D.Miller,
A.Morse,
K.Moy,
J.Ouyang,
R.Page,
A.Robb,
K.Rodrigues,
T.L.Selby,
G.Spraggon,
R.C.Stevens,
H.van den Bedem,
J.Velasquez,
J.Vincent,
X.Wang,
B.West,
G.Wolf,
K.O.Hodgson,
J.Wooley,
I.A.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Crystal structure of TM0920. A: Ribbon diagram of
Thermotoga maritima TM0920 1,3-propanediol dehydrogenase color
coded from N-terminus (blue) to C-terminus (red) showing the
domain organization and location of the active site (arrow).
 -helices
(H1-H19) and -helices
(H1-H19) and  -strands
( -strands
(  1- 1-
 8)
are indicated. The NADP^+ molecule is shown in ball and stick.
The Fe^2+ ion is shown as a sphere in magenta. B: Diagram
showing the secondary structure elements in TM0920 superimposed
on its primary sequence. Residues located in the active site and
interacting with Fe^2+ are indicated by blue dots and green
triangles. Residues interacting with NADP^+ are indicated by red
dots. The location of the 8)
are indicated. The NADP^+ molecule is shown in ball and stick.
The Fe^2+ ion is shown as a sphere in magenta. B: Diagram
showing the secondary structure elements in TM0920 superimposed
on its primary sequence. Residues located in the active site and
interacting with Fe^2+ are indicated by blue dots and green
triangles. Residues interacting with NADP^+ are indicated by red
dots. The location of the  -hairpin
formed by -hairpin
formed by  -strands
6 and 7 is depicted in red. -strands
6 and 7 is depicted in red.
|
 |
Figure 2.
Figure 2. A: Schematic representation of the interactions
between NADP^+ and its interacting residues. The NADP^+ molecule
is depicted in purple and the protein residues are in orange.
The atoms are indicated as follows: carbon (black), oxygen
(red), nitrogen (blue), and phosphorus (purple). Hydrogen bonds
are represented as dashed green lines. Residues implicated in
hydrophobic interaction are represented as barbed circle
sections. B: Close-up view of the active site showing residues
coordinating the metal ion.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2004,
54,
174-177)
copyright 2004.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
L.R.Jarboe
(2011).
YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals.
|
| |
Appl Microbiol Biotechnol,
89,
249-257.
|
 |
|
|
|
|
 |
S.Atsumi,
T.Y.Wu,
E.M.Eckl,
S.D.Hawkins,
T.Buelter,
and
J.C.Liao
(2010).
Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes.
|
| |
Appl Microbiol Biotechnol,
85,
651-657.
|
 |
|
|
|
|
 |
D.Marçal,
A.T.Rêgo,
M.A.Carrondo,
and
F.J.Enguita
(2009).
1,3-Propanediol dehydrogenase from Klebsiella pneumoniae: decameric quaternary structure and possible subunit cooperativity.
|
| |
J Bacteriol,
191,
1143-1151.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
X.Liu,
Y.Dong,
J.Zhang,
A.Zhang,
L.Wang,
and
L.Feng
(2009).
Two novel metal-independent long-chain alkyl alcohol dehydrogenases from Geobacillus thermodenitrificans NG80-2.
|
| |
Microbiology,
155,
2078-2085.
|
 |
|
|
|
|
 |
X.Ying,
A.M.Grunden,
L.Nie,
M.W.Adams,
and
K.Ma
(2009).
Molecular characterization of the recombinant iron-containing alcohol dehydrogenase from the hyperthermophilic Archaeon, Thermococcus strain ES1.
|
| |
Extremophiles,
13,
299-311.
|
 |
|
|
|
|
 |
X.Ying,
Y.Wang,
H.R.Badiei,
V.Karanassios,
and
K.Ma
(2007).
Purification and characterization of an iron-containing alcohol dehydrogenase in extremely thermophilic bacterium Thermotoga hypogea.
|
| |
Arch Microbiol,
187,
499-510.
|
 |
|
|
|
|
 |
R.Sparling,
R.Islam,
N.Cicek,
C.Carere,
H.Chow,
and
D.B.Levin
(2006).
Formate synthesis by Clostridium thermocellum during anaerobic fermentation.
|
| |
Can J Microbiol,
52,
681-688.
|
 |
|
|
|
|
 |
C.Montella,
L.Bellsolell,
R.Pérez-Luque,
J.Badía,
L.Baldoma,
M.Coll,
and
J.Aguilar
(2005).
Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli.
|
| |
J Bacteriol,
187,
4957-4966.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.Page,
A.M.Deacon,
S.A.Lesley,
and
R.C.Stevens
(2005).
Shotgun crystallization strategy for structural genomics II: crystallization conditions that produce high resolution structures for T. maritima proteins.
|
| |
J Struct Funct Genomics,
6,
209-217.
|
 |
|
|
|
|
 |
P.H.Zwart,
G.G.Langer,
and
V.S.Lamzin
(2004).
Modelling bound ligands in protein crystal structures.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
2230-2239.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

