 |
PDBsum entry 1nyl

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.18
- glutamine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Gln) + L-glutamine + ATP = L-glutaminyl-tRNA(Gln) + AMP + diphosphate
|
 |
 |
 |
 |
 |
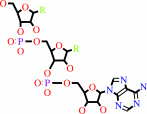 tRNA(Gln)
tRNA(Gln)
|
+
|
 L-glutamine
L-glutamine
|
+
|
 ATP
ATP
|
=
|
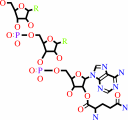 L-glutaminyl-tRNA(Gln)
L-glutaminyl-tRNA(Gln)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Structure
11:591-603
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
tRNA-dependent active site assembly in a class I aminoacyl-tRNA synthetase.
|
|
L.D.Sherlin,
J.J.Perona.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of ligand-free E. coli glutaminyl-tRNA synthetase (GlnRS)
at 2.4 A resolution shows that substrate binding is essential to construction of
a catalytically proficient active site. tRNA binding generates structural
changes throughout the enzyme, repositioning key active site peptides that bind
glutamine and ATP. The structure gives insight into longstanding questions
regarding the tRNA dependence of glutaminyl adenylate formation, the coupling of
amino acid and tRNA selectivities, and the roles of specific pathways for
transmission of tRNA binding signals to the active site. Comparative analysis of
the unliganded and tRNA-bound structures shows, in detail, how flexibility is
built into the enzyme architecture and suggests that the induced-fit transitions
are a key underlying determinant of both amino acid and tRNA specificity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 5.
Figure 5. Substrate Juxtaposition by Induced Fit(A) Stereo
view of enzyme conformational changes in the vicinity of the
tRNA 3'-end. The tRNA-bound and unliganded enzymes are
superimposed on the basis of equivalent residues in the DNF
domains (see text). tRNA, dark blue; unliganded enzyme, light
blue; complexed enzyme, gray (DNF domain) and pink (ABD). Dotted
red lines indicate hydrogen bonds observed in the GlnRS-tRNA
complex. QSI inhibitor from the complexed structure, red.(B)
Stereo view of enzyme conformational changes in the active site,
with structures superimposed as in (A). Complexed enzyme, gray;
unliganded enzyme, blue; QSI inhibitor, red. The disorder in the
surface loop at Val71 in the unliganded enzyme is shown by the
dotted blue line (right). Dotted red lines indicate hydrogen
bonds observed in the GlnRS-tRNA complex.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from Cell Press:
Structure
(2003,
11,
591-603)
copyright 2003.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Rodríguez-Hernández,
and
J.J.Perona
(2011).
Heat maps for intramolecular communication in an RNP enzyme encoding glutamine.
|
| |
Structure,
19,
386-396.
|
 |
|
|
|
|
 |
E.M.Corigliano,
and
J.J.Perona
(2009).
Architectural underpinnings of the genetic code for glutamine.
|
| |
Biochemistry,
48,
676-687.
|
 |
|
|
|
|
 |
G.L.Igloi,
and
E.Schiefermayr
(2009).
Amino acid discrimination by arginyl-tRNA synthetases as revealed by an examination of natural specificity variants.
|
| |
FEBS J,
276,
1307-1318.
|
 |
|
|
|
|
 |
C.M.Zhang,
C.Liu,
T.Christian,
H.Gamper,
J.Rozenski,
D.Pan,
J.B.Randolph,
E.Wickstrom,
B.S.Cooperman,
and
Y.M.Hou
(2008).
Pyrrolo-C as a molecular probe for monitoring conformations of the tRNA 3' end.
|
| |
RNA,
14,
2245-2253.
|
 |
|
|
|
|
 |
C.S.Francklyn
(2008).
DNA polymerases and aminoacyl-tRNA synthetases: shared mechanisms for ensuring the fidelity of gene expression.
|
| |
Biochemistry,
47,
11695-11703.
|
 |
|
|
|
|
 |
T.L.Bullock,
A.Rodríguez-Hernández,
E.M.Corigliano,
and
J.J.Perona
(2008).
A rationally engineered misacylating aminoacyl-tRNA synthetase.
|
| |
Proc Natl Acad Sci U S A,
105,
7428-7433.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Deniziak,
C.Sauter,
H.D.Becker,
C.A.Paulus,
R.Giegé,
and
D.Kern
(2007).
Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation.
|
| |
Nucleic Acids Res,
35,
1421-1431.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.E.Budiman,
M.H.Knaggs,
J.S.Fetrow,
and
R.W.Alexander
(2007).
Using molecular dynamics to map interaction networks in an aminoacyl-tRNA synthetase.
|
| |
Proteins,
68,
670-689.
|
 |
|
|
|
|
 |
M.Kapustina,
V.Weinreb,
L.Li,
B.Kuhlman,
and
C.W.Carter
(2007).
A conformational transition state accompanies tryptophan activation by B. stearothermophilus tryptophanyl-tRNA synthetase.
|
| |
Structure,
15,
1272-1284.
|
 |
|
|
|
|
 |
R.Sathyapriya,
and
S.Vishveshwara
(2007).
Structure networks of E. coli glutaminyl-tRNA synthetase: effects of ligand binding.
|
| |
Proteins,
68,
541-550.
|
 |
|
|
|
|
 |
S.W.Lue,
and
S.O.Kelley
(2007).
A single residue in leucyl-tRNA synthetase affecting amino acid specificity and tRNA aminoacylation.
|
| |
Biochemistry,
46,
4466-4472.
|
 |
|
|
|
|
 |
N.T.Uter,
and
J.J.Perona
(2006).
Active-site assembly in glutaminyl-tRNA synthetase by tRNA-mediated induced fit.
|
| |
Biochemistry,
45,
6858-6865.
|
 |
|
|
|
|
 |
I.Gruic-Sovulj,
N.Uter,
T.Bullock,
and
J.J.Perona
(2005).
tRNA-dependent aminoacyl-adenylate hydrolysis by a nonediting class I aminoacyl-tRNA synthetase.
|
| |
J Biol Chem,
280,
23978-23986.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.T.Uter,
I.Gruic-Sovulj,
and
J.J.Perona
(2005).
Amino acid-dependent transfer RNA affinity in a class I aminoacyl-tRNA synthetase.
|
| |
J Biol Chem,
280,
23966-23977.
|
 |
|
|
|
|
 |
R.Powers,
N.Mirkovic,
S.Goldsmith-Fischman,
T.B.Acton,
Y.Chiang,
Y.J.Huang,
L.Ma,
P.K.Rajan,
J.R.Cort,
M.A.Kennedy,
J.Liu,
B.Rost,
B.Honig,
D.Murray,
and
G.T.Montelione
(2005).
Solution structure of Archaeglobus fulgidis peptidyl-tRNA hydrolase (Pth2) provides evidence for an extensive conserved family of Pth2 enzymes in archea, bacteria, and eukaryotes.
|
| |
Protein Sci,
14,
2849-2861.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
J.Levengood,
S.F.Ataide,
H.Roy,
and
M.Ibba
(2004).
Divergence in noncognate amino acid recognition between class I and class II lysyl-tRNA synthetases.
|
| |
J Biol Chem,
279,
17707-17714.
|
 |
|
|
|
|
 |
S.Hauenstein,
C.M.Zhang,
Y.M.Hou,
and
J.J.Perona
(2004).
Shape-selective RNA recognition by cysteinyl-tRNA synthetase.
|
| |
Nat Struct Mol Biol,
11,
1134-1141.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
W.H.McClain,
K.Gabriel,
D.Lee,
and
S.Otten
(2004).
Structure-function analysis of tRNA(Gln) in an Escherichia coli knockout strain.
|
| |
RNA,
10,
795-804.
|
 |
|
|
|
|
 |
J.Cavarelli
(2003).
Pushing induced fit to its limits: tRNA-dependent active site assembly in class I aminoacyl-tRNA synthetases.
|
| |
Structure,
11,
484-486.
|
 |
|
|
|
|
 |
R.Banerjee,
A.K.Mandal,
R.Saha,
S.Guha,
S.Samaddar,
A.Bhattacharyya,
and
S.Roy
(2003).
Solvation change and ion release during aminoacylation by aminoacyl-tRNA synthetases.
|
| |
Nucleic Acids Res,
31,
6035-6042.
|
 |
|
|
|
|
 |
R.Geslain,
G.Bey,
J.Cavarelli,
and
G.Eriani
(2003).
Limited set of amino acid residues in a class Ia aminoacyl-tRNA synthetase is crucial for tRNA binding.
|
| |
Biochemistry,
42,
15092-15101.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

