 |
PDBsum entry 1nsm

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.1.3.3
- aldose 1-epimerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
alpha-D-glucose = beta-D-glucose
|
 |
 |
 |
 |
 |
alpha-D-glucose
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
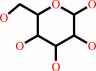 beta-D-glucose
beta-D-glucose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
12:1051-1059
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
The catalytic mechanism of galactose mutarotase.
|
|
J.B.Thoden,
J.Kim,
F.M.Raushel,
H.M.Holden.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Galactose mutarotase catalyzes the first step in normal galactose metabolism by
catalyzing the conversion of beta-D-galactose to alpha-D-galactose. The
structure of the enzyme from Lactococcus lactis was recently solved in this
laboratory and shown to be topologically similar to domain 5 of
beta-galactosidase. From this initial X-ray analysis, four amino acid residues
were demonstrated to be intimately involved in sugar binding to the protein: His
96, His 170, Asp 243, and Glu 304. Here we present a combined X-ray
crystallographic and kinetic analysis designed to examine the role of these
residues in the reaction mechanism of the enzyme. For this investigation, the
following site-directed mutant proteins were prepared: H96N, H170N, D243N,
D243A, E304Q, and E304A. All of the structures of these proteins, complexed with
either glucose or galactose, were solved to a nominal resolution of 1.95 A or
better, and their kinetic parameters were measured against D-galactose,
D-glucose, L-arabinose, or D-xylose. From these studies, it can be concluded
that Glu 304 and His 170 are critical for catalysis and that His 96 and Asp 243
are important for proper substrate positioning within the active site.
Specifically, Glu 304 serves as the active site base to initiate the reaction by
removing the proton from the C-1 hydroxyl group of the sugar substrate and His
170 functions as the active site acid to protonate the C-5 ring oxygen.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Active site of galactose mutarotase from L.
lactis. A close-up view of the active site within ~5 Å of the
galactose ligand is displayed in (A). Ordered water molecules
surrounding the sugar ligand were omitted for figure clarity.
Both the  - and ß-anomers
of galactose were observed in the electron density map. A
close-up view of the active site within ~5 Å of bound glucose is
presented in (B). All figures in this article were prepared with
the software package MOLSCRIPT (Kraulis 1991). - and ß-anomers
of galactose were observed in the electron density map. A
close-up view of the active site within ~5 Å of bound glucose is
presented in (B). All figures in this article were prepared with
the software package MOLSCRIPT (Kraulis 1991).
|
 |
Figure 3.
Figure 3. Schematics of hydrogen bonding patterns. Shown in
(A) is the observed hydrogen bonding pattern between the
wild-type protein and galactose. Both the  - and ß-anomers
at C-1 are observed in the active site. The hydrogen bonding
pattern exhibited between the E304Q mutant protein and glucose
is depicted in (B). - and ß-anomers
at C-1 are observed in the active site. The hydrogen bonding
pattern exhibited between the E304Q mutant protein and glucose
is depicted in (B).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2003,
12,
1051-1059)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Yang,
S.Park,
L.Makowski,
and
B.Roux
(2009).
A rapid coarse residue-based computational method for x-ray solution scattering characterization of protein folds and multiple conformational states of large protein complexes.
|
| |
Biophys J,
96,
4449-4463.
|
 |
|
|
|
|
 |
J.S.Richardson,
X.Carpena,
J.Switala,
R.Perez-Luque,
L.J.Donald,
P.C.Loewen,
and
I.J.Oresnik
(2008).
RhaU of Rhizobium leguminosarum is a rhamnose mutarotase.
|
| |
J Bacteriol,
190,
2903-2910.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.J.Lee,
D.E.Lewis,
and
S.Adhya
(2008).
Induction of the galactose enzymes in Escherichia coli is independent of the C-1-hydroxyl optical configuration of the inducer D-galactose.
|
| |
J Bacteriol,
190,
7932-7938.
|
 |
|
|
|
|
 |
S.Chittori,
D.K.Simanshu,
H.S.Savithri,
and
M.R.Murthy
(2007).
Structure of the putative mutarotase YeaD from Salmonella typhimurium: structural comparison with galactose mutarotases.
|
| |
Acta Crystallogr D Biol Crystallogr,
63,
197-205.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Assairi,
T.Bertrand,
J.Ferdinand,
N.Slavova-Azmanova,
M.Christensen,
P.Briozzo,
F.Schaeffer,
C.T.Craescu,
J.Neuhard,
O.Bârzu,
and
A.M.Gilles
(2004).
Deciphering the function of an ORF: Salmonella enterica DeoM protein is a new mutarotase specific for deoxyribose.
|
| |
Protein Sci,
13,
1295-1303.
|
 |
|
|
|
|
 |
S.Majumdar,
J.Ghatak,
S.Mukherji,
H.Bhattacharjee,
and
A.Bhaduri
(2004).
UDPgalactose 4-epimerase from Saccharomyces cerevisiae. A bifunctional enzyme with aldose 1-epimerase activity.
|
| |
Eur J Biochem,
271,
753-759.
|
 |
|
|
|
|
 |
T.Vilfan,
B.Cresnar,
D.Fournier,
J.Stojan,
and
K.Breskvar
(2004).
Characterisation and expression of a gene encoding a mutarotase from the fungus Rhizopus nigricans.
|
| |
FEMS Microbiol Lett,
235,
101-108.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

