 |
PDBsum entry 1njs

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Human gar tfase in complex with hydrolyzed form of 10-trifluoroacetyl- 5,10-dideaza-acyclic-5,6,7,8-tetrahydrofolic acid
|
|
Structure:
|
 |
Phosphoribosylglycinamide formyltransferase. Chain: a, b. Synonym: gar tfase, gart, gar transformylase, 5'- phosphoribosylglycinamide transformylase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: gart. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
1.98Å
|
R-factor:
|
0.228
|
R-free:
|
0.247
|
|
|
Authors:
|
 |
Y.Zhang,J.Desharnais,T.H.Marsilje,C.Li,M.P.Hedrick,L.T.Gooljarsingh, A.Tavassoli,S.J.Benkovic,A.J.Olson,D.L.Boger,I.A.Wilson
|
Key ref:

|
 |
Y.Zhang
et al.
(2003).
Rational design, synthesis, evaluation, and crystal structure of a potent inhibitor of human GAR Tfase: 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid.
Biochemistry,
42,
6043-6056.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
02-Jan-03
|
Release date:
|
10-Jun-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P22102
(PUR2_HUMAN) -
Trifunctional purine biosynthetic protein adenosine-3 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1010 a.a.
200 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.1.2.2
- phosphoribosylglycinamide formyltransferase 1.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Purine Biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N1-(5-phospho-beta-D-ribosyl)glycinamide + (6R)-10- formyltetrahydrofolate = N2-formyl-N1-(5-phospho-beta-D- ribosyl)glycinamide + (6S)-5,6,7,8-tetrahydrofolate + H+
|
 |
 |
 |
 |
 |
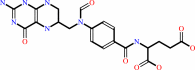 10-formyltetrahydrofolate
10-formyltetrahydrofolate
|
+
|
N(1)-(5-phospho-D-ribosyl)glycinamide
|
=
|
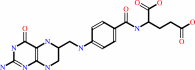 tetrahydrofolate
tetrahydrofolate
|
+
|
N(2)-formyl-N(1)-(5-phospho-D-ribosyl)glycinamide
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.6.3.3.1
- phosphoribosylformylglycinamidine cyclo-ligase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-formamido-N1-(5-O-phospho-beta-D-ribosyl)acetamidine + ATP = 5-amino- 1-(5-phospho-beta-D-ribosyl)imidazole + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
2-formamido-N(1)-(5-O-phospho-beta-D-ribosyl)acetamidine
|
+
|
 ATP
ATP
|
=
|
5-amino- 1-(5-phospho-beta-D-ribosyl)imidazole
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Magnesium
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.6.3.4.13
- phosphoribosylamine--glycine ligase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-phospho-beta-D-ribosylamine + glycine + ATP = N1-(5-phospho-beta-D- ribosyl)glycinamide + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
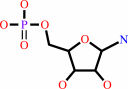 5-phospho-beta-D-ribosylamine
5-phospho-beta-D-ribosylamine
|
+
|
 glycine
glycine
|
+
|
 ATP
ATP
|
=
|
N(1)-(5-phospho-beta-D- ribosyl)glycinamide
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
42:6043-6056
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Rational design, synthesis, evaluation, and crystal structure of a potent inhibitor of human GAR Tfase: 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid.
|
|
Y.Zhang,
J.Desharnais,
T.H.Marsilje,
C.Li,
M.P.Hedrick,
L.T.Gooljarsingh,
A.Tavassoli,
S.J.Benkovic,
A.J.Olson,
D.L.Boger,
I.A.Wilson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Glycinamide ribonucleotide transformylase (GAR Tfase) has been the target of
anti-neoplastic intervention for almost two decades. Here, we use a
structure-based approach to design a novel folate analogue,
10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid
(10-CF(3)CO-DDACTHF, 1), which specifically inhibits recombinant human GAR Tfase
(K(i) = 15 nM), but is inactive (K(i) > 100 microM) against other
folate-dependent enzymes that have been examined. Moreover, compound 1 is a
potent inhibitor of tumor cell proliferation (IC(50) = 16 nM, CCRF-CEM), which
represents a 10-fold improvement over Lometrexol, a GAR Tfase inhibitor that has
been in clinical trials. Thus, this folate analogue 1 is among the most potent
and selective inhibitors known toward GAR Tfase. Contributing to its efficacious
activity, compound 1 is effectively transported into the cell by the reduced
folate carrier and intracellularly sequestered by polyglutamation. The crystal
structure of human GAR Tfase with folate analogue 1 at 1.98 A resolution
represents the first structure of any GAR Tfase to be determined with a cofactor
or cofactor analogue without the presence of substrate. The folate-binding loop
of residues 141-146, which is highly flexible in both Escherichia coli and
unliganded human GAR Tfase structures, becomes highly ordered upon binding 1 in
the folate-binding site. Computational docking of the natural cofactor into this
and other apo or complexed structures provides a rational basis for modeling how
the natural cofactor 10-formyltetrahydrofolic acid interacts with GAR Tfase, and
suggests that this folate analogue-bound conformation represents the best
template to date for inhibitor design.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.M.Murta,
T.J.Vickers,
D.A.Scott,
and
S.M.Beverley
(2009).
Methylene tetrahydrofolate dehydrogenase/cyclohydrolase and the synthesis of 10-CHO-THF are essential in Leishmania major.
|
| |
Mol Microbiol,
71,
1386-1401.
|
 |
|
|
|
|
 |
J.K.DeMartino,
I.Hwang,
S.Connelly,
I.A.Wilson,
and
D.L.Boger
(2008).
Asymmetric synthesis of inhibitors of glycinamide ribonucleotide transformylase.
|
| |
J Med Chem,
51,
5441-5448.
|
 |
|
|
|
|
 |
Y.Zhang,
M.Morar,
and
S.E.Ealick
(2008).
Structural biology of the purine biosynthetic pathway.
|
| |
Cell Mol Life Sci,
65,
3699-3724.
|
 |
|
|
|
|
 |
W.Manieri,
M.E.Moore,
M.B.Soellner,
P.Tsang,
and
C.A.Caperelli
(2007).
Human glycinamide ribonucleotide transformylase: active site mutants as mechanistic probes.
|
| |
Biochemistry,
46,
156-163.
|
 |
|
|
|
|
 |
Y.G.Assaraf
(2007).
Molecular basis of antifolate resistance.
|
| |
Cancer Metastasis Rev,
26,
153-181.
|
 |
|
|
|
|
 |
J.K.DeMartino,
I.Hwang,
L.Xu,
I.A.Wilson,
and
D.L.Boger
(2006).
Discovery of a potent, nonpolyglutamatable inhibitor of glycinamide ribonucleotide transformylase.
|
| |
J Med Chem,
49,
2998-3002.
|
 |
|
|
|
|
 |
Y.G.Assaraf
(2006).
The role of multidrug resistance efflux transporters in antifolate resistance and folate homeostasis.
|
| |
Drug Resist Updat,
9,
227-246.
|
 |
|
|
|
|
 |
P.Z.Gatzeva-Topalova,
A.P.May,
and
M.C.Sousa
(2005).
Crystal structure and mechanism of the Escherichia coli ArnA (PmrI) transformylase domain. An enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance.
|
| |
Biochemistry,
44,
5328-5338.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
C.G.Cheong,
D.W.Wolan,
S.E.Greasley,
P.A.Horton,
G.P.Beardsley,
and
I.A.Wilson
(2004).
Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates.
|
| |
J Biol Chem,
279,
18034-18045.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Xu,
C.Li,
A.J.Olson,
and
I.A.Wilson
(2004).
Crystal structure of avian aminoimidazole-4-carboxamide ribonucleotide transformylase in complex with a novel non-folate inhibitor identified by virtual ligand screening.
|
| |
J Biol Chem,
279,
50555-50565.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

