 |
PDBsum entry 1n2h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.1
- pantoate--beta-alanine ligase (AMP-forming).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Coenzyme A Biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-pantoate + beta-alanine + ATP = (R)-pantothenate + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
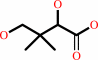 (R)-pantoate
(R)-pantoate
|
+
|
 beta-alanine
beta-alanine
|
+
|
ATP
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
=
|
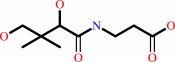 (R)-pantothenate
(R)-pantothenate
|
+
|
AMP
Bound ligand (Het Group name = )
matches with 71.88% similarity
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Protein Sci
12:1097-1108
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of a pantothenate synthetase from M. tuberculosis and its complexes with substrates and a reaction intermediate.
|
|
S.Wang,
D.Eisenberg.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Pantothenate biosynthesis is essential for the virulence of Mycobacterium
tuberculosis, and this pathway thus presents potential drug targets against
tuberculosis. We determined the crystal structure of pantothenate synthetase
(PS) from M. tuberculosis, and its complexes with AMPCPP, pantoate, and a
reaction intermediate, pantoyl adenylate, with resolutions from 1.6 to 2 A. PS
catalyzes the ATP-dependent condensation of pantoate and beta-alanine to form
pantothenate. Its structure reveals a dimer, and each subunit has two domains
with tight association between domains. The active-site cavity is on the
N-terminal domain, partially covered by the C-terminal domain. One wall of the
active site cavity is flexible, which allows the bulky AMPCPP to diffuse into
the active site to nearly full occupancy when crystals are soaked in solutions
containing AMPCPP. Crystal structures of the complexes with AMPCPP and pantoate
indicate that the enzyme binds ATP and pantoate tightly in the active site, and
brings the carboxyl oxygen of pantoate near the alpha-phosphorus atom of ATP for
an in-line nucleophilic attack. When crystals were soaked with, or grown in the
presence of, both ATP and pantoate, a reaction intermediate, pantoyl adenylate,
is found in the active site. The flexible wall of the active site cavity becomes
ordered when the intermediate is in the active site, thus protecting it from
being hydrolyzed. Binding of beta-alanine can occur only after pantoyl adenylate
is formed inside the active site cavity. The tight binding of the intermediate
pantoyl adenylate suggests that nonreactive analogs of pantoyl adenylate may be
inhibitors of the PS enzyme with high affinity and specificity.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Ribbon diagram of the M. tuberculosis
pantothenate synthetase dimer. (A) A side view of the dimer
structure showing that it resembles the shape of a butterfly.
(B) An orthogonal view of (A) from top, with the twofold NCS
symmetry axis (labeled with a dot) approximately perpendicular
to the paper plane. Secondary structure elements for the subunit
A (left) are labeled. Those for subunit B are identical except
that the short helix  3' is not
present. The figure was prepared from the coordinates of the
intermediate complex (data set 5), with the program Molscript
(Kraulis 1991) and Raster3D (Merritt and Murphy 1994). The
molecule in the active site of each subunit, shown in
ball-and-stick, is the reaction intermediate, pantoyl adenylate. 3' is not
present. The figure was prepared from the coordinates of the
intermediate complex (data set 5), with the program Molscript
(Kraulis 1991) and Raster3D (Merritt and Murphy 1994). The
molecule in the active site of each subunit, shown in
ball-and-stick, is the reaction intermediate, pantoyl adenylate.
|
 |
Figure 4.
Figure 4. Active site cavity and the binding of AMPCPP,
pantoate, and pantoyl adenylate. (A) A stereo view of the active
site cavity of subunit A of the complex with both AMPCPP and
pantoate. The substrates (both with partial occupancy) are shown
as ball-and-stick models. The active site cavity is surrounded
by ß2-loop-  2, ß7-loop,
ß6-loop- 2, ß7-loop,
ß6-loop-  6,
3[10]5'-loop- 6,
3[10]5'-loop-  5, and
ß3-loop-3[10]3- 5, and
ß3-loop-3[10]3-  3'-loop, and
covered by 3[10]7 and the ß-sheet of C-terminal domain. Residues
around helix 3'-loop, and
covered by 3[10]7 and the ß-sheet of C-terminal domain. Residues
around helix  3' (shown in
cyan) are disordered in subunit B, which has a fully occupied
AMPCPP and a glycerol molecule in the active site. (B) A section
of the initial difference electron density map (Fo - Fc) in the
active site of subunit B superimposed on the refined model,
calculated at 1.7 Å and contoured at the 2 3' (shown in
cyan) are disordered in subunit B, which has a fully occupied
AMPCPP and a glycerol molecule in the active site. (B) A section
of the initial difference electron density map (Fo - Fc) in the
active site of subunit B superimposed on the refined model,
calculated at 1.7 Å and contoured at the 2  level. Side
chains of Lys160, Ser196, and Arg198 have moved relative to
those in the apo enzyme to interact with the phosphate groups,
and thus also have positive initial difference electron density.
The electron density figures are prepared with PYMOL (DeLano
2002). (C) Detailed binding interactions between AMPCPP (shown
with carbon atoms in gold) and protein active site residues of
subunit B. The Mg2+ ion is shown as a yellow sphere, and water
molecules are shown as red spheres. Hydrogen bonds between
AMPCPP and protein atoms, and some water-mediated hydrogen bonds
are shown as dashed lines. A glycerol molecule found next to the level. Side
chains of Lys160, Ser196, and Arg198 have moved relative to
those in the apo enzyme to interact with the phosphate groups,
and thus also have positive initial difference electron density.
The electron density figures are prepared with PYMOL (DeLano
2002). (C) Detailed binding interactions between AMPCPP (shown
with carbon atoms in gold) and protein active site residues of
subunit B. The Mg2+ ion is shown as a yellow sphere, and water
molecules are shown as red spheres. Hydrogen bonds between
AMPCPP and protein atoms, and some water-mediated hydrogen bonds
are shown as dashed lines. A glycerol molecule found next to the
 -phosphate of
AMPCPP, at the pantoate binding site, is also shown. (D) A
section of the initial difference electron density (Fo - Fc)
around the bound pantoate molecule in the active site of subunit
A of the pantoate-ß-alanine complex (data set 7) shows that
pantoate is very well ordered with full occupancy. The nearby
residues did not move relative to those of the apo enzyme, and
therefore did not have initial difference density. The electron
density was calculated at 1.7 Å and contoured at 2 -phosphate of
AMPCPP, at the pantoate binding site, is also shown. (D) A
section of the initial difference electron density (Fo - Fc)
around the bound pantoate molecule in the active site of subunit
A of the pantoate-ß-alanine complex (data set 7) shows that
pantoate is very well ordered with full occupancy. The nearby
residues did not move relative to those of the apo enzyme, and
therefore did not have initial difference density. The electron
density was calculated at 1.7 Å and contoured at 2  . (E) The
pantoate molecule (shown in gold for the carbon atoms) is
tightly bound and fits snugly in its binding site. Two glutamine
side chains form hydrogen bonds to the hydroxyl groups and one
carboxyl oxygen of the pantoate. The two methyl groups and the
hydrophobic side of pantoate interact with the side chains of
Pro38, Met40, and Phe157. (F) A section of the initial
difference electron density (Fo - Fc) around the bound pantoyl
adenylate molecule in the active site of subunit B of the
intermediate complex (data set 6) shows that intermediate is
very well ordered with full occupancy. The electron density was
calculated at 1.7 Å and contoured at 2 . (E) The
pantoate molecule (shown in gold for the carbon atoms) is
tightly bound and fits snugly in its binding site. Two glutamine
side chains form hydrogen bonds to the hydroxyl groups and one
carboxyl oxygen of the pantoate. The two methyl groups and the
hydrophobic side of pantoate interact with the side chains of
Pro38, Met40, and Phe157. (F) A section of the initial
difference electron density (Fo - Fc) around the bound pantoyl
adenylate molecule in the active site of subunit B of the
intermediate complex (data set 6) shows that intermediate is
very well ordered with full occupancy. The electron density was
calculated at 1.7 Å and contoured at 2  . (G) The
pantoyl adenylate molecule (shown with carbon atoms in gold) is
tightly bound and fits snugly in the active site cavity. The
adenosine and pantoyl groups are at identical positions as those
in the AMPCPP complex and pantoate complex, respectively, and
have identical interactions with the active site residues.
However, the . (G) The
pantoyl adenylate molecule (shown with carbon atoms in gold) is
tightly bound and fits snugly in the active site cavity. The
adenosine and pantoyl groups are at identical positions as those
in the AMPCPP complex and pantoate complex, respectively, and
have identical interactions with the active site residues.
However, the  -phosphate
moved down to have a covalent bond to the pantoate, which allows
the phosphate group to have a hydrogen bond to the amide
nitrogen of Met40. -phosphate
moved down to have a covalent bond to the pantoate, which allows
the phosphate group to have a hydrogen bond to the amide
nitrogen of Met40.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the Protein Society:
Protein Sci
(2003,
12,
1097-1108)
copyright 2003.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.S.Tan,
G.Fuentes,
and
C.Verma
(2011).
A comparison of the dynamics of pantothenate synthetase from M. tuberculosis and E. coli: Computational studies.
|
| |
Proteins,
79,
1715-1727.
|
 |
|
|
|
|
 |
K.S.Chakrabarti,
K.G.Thakur,
B.Gopal,
and
S.P.Sarma
(2010).
X-ray crystallographic and NMR studies of pantothenate synthetase provide insights into the mechanism of homotropic inhibition by pantoate.
|
| |
FEBS J,
277,
697-712.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.E.Scott,
G.J.Dawes,
M.Ando,
C.Abell,
and
A.Ciulli
(2009).
A fragment-based approach to probing adenosine recognition sites by using dynamic combinatorial chemistry.
|
| |
Chembiochem,
10,
2772-2779.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
T.R.Ioerger,
and
J.C.Sacchettini
(2009).
Structural genomics approach to drug discovery for Mycobacterium tuberculosis.
|
| |
Curr Opin Microbiol,
12,
318-325.
|
 |
|
|
|
|
 |
A.Ciulli,
D.E.Scott,
M.Ando,
F.Reyes,
S.A.Saldanha,
K.L.Tuck,
D.Y.Chirgadze,
T.L.Blundell,
and
C.Abell
(2008).
Inhibition of Mycobacterium tuberculosis pantothenate synthetase by analogues of the reaction intermediate.
|
| |
Chembiochem,
9,
2606-2611.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Seetharamappa,
M.Oke,
H.Liu,
S.A.McMahon,
K.A.Johnson,
L.Carter,
M.Dorward,
M.Zawadzki,
I.M.Overton,
C.A.van Niekirk,
S.Graham,
C.H.Botting,
G.L.Taylor,
M.F.White,
G.J.Barton,
P.J.Coote,
and
J.H.Naismith
(2007).
Purification, crystallization and data collection of methicillin-resistant Staphylococcus aureus Sar2676, a pantothenate synthetase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
488-491.
|
 |
|
|
|
|
 |
K.L.Tuck,
S.A.Saldanha,
L.M.Birch,
A.G.Smith,
and
C.Abell
(2006).
The design and synthesis of inhibitors of pantothenate synthetase.
|
| |
Org Biomol Chem,
4,
3598-3610.
|
 |
|
|
|
|
 |
V.L.Arcus,
J.S.Lott,
J.M.Johnston,
and
E.N.Baker
(2006).
The potential impact of structural genomics on tuberculosis drug discovery.
|
| |
Drug Discov Today,
11,
28-34.
|
 |
|
|
|
|
 |
J.Minshull,
J.E.Ness,
C.Gustafsson,
and
S.Govindarajan
(2005).
Predicting enzyme function from protein sequence.
|
| |
Curr Opin Chem Biol,
9,
202-209.
|
 |
|
|
|
|
 |
C.V.Smith,
and
J.C.Sacchettini
(2003).
Mycobacterium tuberculosis: a model system for structural genomics.
|
| |
Curr Opin Struct Biol,
13,
658-664.
|
 |
|
|
|
|
 |
M.Bellinzoni,
and
G.Riccardi
(2003).
Techniques and applications: The heterologous expression of Mycobacterium tuberculosis genes is an uphill road.
|
| |
Trends Microbiol,
11,
351-358.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

