 |
PDBsum entry 1m3e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.8.3.5
- 3-oxoacid CoA-transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 3-oxo acid + succinyl-CoA = a 3-oxoacyl-CoA + succinate
|
 |
 |
 |
 |
 |
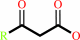 3-oxo acid
3-oxo acid
|
+
|
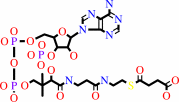 succinyl-CoA
succinyl-CoA
|
=
|
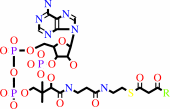 3-oxoacyl-CoA
3-oxoacyl-CoA
|
+
|
 succinate
succinate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:14455-14462
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of the mammalian CoA transferase from pig heart.
|
|
K.S.Bateman,
E.R.Brownie,
W.T.Wolodko,
M.E.Fraser.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ketoacidosis affects patients who are deficient in the enzyme activity of
succinyl-CoA:3-ketoacid CoA transferase (SCOT), since SCOT catalyses the
activation of acetoacetate in the metabolism of ketone bodies. Thus far,
structure/function analysis of the mammalian enzyme has been predicted based on
the three-dimensional structure of a CoA transferase determined from an
anaerobic bacterium that utilizes its enzyme for glutamate fermentation. To
better interpret clinical data, we have determined the structure of a mammalian
CoA transferase from pig heart by X-ray crystallography to 2.5 A resolution.
Instrumental to the structure determination were selenomethionine substitution
and the use of argon during purification and crystallization. Although pig heart
SCOT adopts an alpha/beta protein fold, resembling the overall fold of the
bacterial CoA transferase, several loops near the active site of pig heart SCOT
follow different paths than the corresponding loops in the bacterial enzyme,
accounting for differences in substrate specificities. Two missense mutations
found associated with SCOT of ketoacidosis patients were mapped to a location in
the structure that might disrupt the stabilization of the amino-terminal strand
and thereby interfere with the proper folding of the protein into a functional
enzyme.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Wang,
H.Sun,
Y.Ru,
S.Yin,
L.Yin,
and
S.Liu
(2011).
Proteomic survey towards the tissue-specific proteins of mouse mitochondria.
|
| |
Sci China Life Sci,
54,
3.
|
 |
|
|
|
|
 |
S.F.Coker,
A.J.Lloyd,
E.Mitchell,
G.R.Lewis,
A.R.Coker,
and
P.M.Shoolingin-Jordan
(2010).
The high-resolution structure of pig heart succinyl-CoA:3-oxoacid coenzyme A transferase.
|
| |
Acta Crystallogr D Biol Crystallogr,
66,
797-805.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Macieira,
J.Zhang,
M.Velarde,
W.Buckel,
and
A.Messerschmidt
(2009).
Crystal structure of 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum.
|
| |
Biol Chem,
390,
1251-1263.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
I.Rebrin,
C.Brégère,
S.Kamzalov,
T.K.Gallaher,
and
R.S.Sohal
(2007).
Nitration of tryptophan 372 in succinyl-CoA:3-ketoacid CoA transferase during aging in rat heart mitochondria.
|
| |
Biochemistry,
46,
10130-10144.
|
 |
|
|
|
|
 |
E.S.Rangarajan,
Y.Li,
E.Ajamian,
P.Iannuzzi,
S.D.Kernaghan,
M.E.Fraser,
M.Cygler,
and
A.Matte
(2005).
Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
|
| |
J Biol Chem,
280,
42919-42928.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.M.Coros,
L.Swenson,
W.T.Wolodko,
and
M.E.Fraser
(2004).
Structure of the CoA transferase from pig heart to 1.7 A resolution.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1717-1725.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Rivière,
S.W.van Weelden,
P.Glass,
P.Vegh,
V.Coustou,
M.Biran,
J.J.van Hellemond,
F.Bringaud,
A.G.Tielens,
and
M.Boshart
(2004).
Acetyl:succinate CoA-transferase in procyclic Trypanosoma brucei. Gene identification and role in carbohydrate metabolism.
|
| |
J Biol Chem,
279,
45337-45346.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

