 |
PDBsum entry 1ld3

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of b. Subilis ferrochelatase with zn(2+) bound at the active site.
|
|
Structure:
|
 |
Ferrochelatase. Chain: a. Engineered: yes
|
|
Source:
|
 |
Bacillus subtilis. Organism_taxid: 1423. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.217
|
R-free:
|
0.274
|
|
|
Authors:
|
 |
D.Lecerof,M.N.Fodje,R.A.Leon,U.Olsson,A.Hansson,E.Sigfridsson,U.Ryde, M.Hansson,S.Al-Karadaghi
|
|
Key ref:
|
 |
D.Lecerof
et al.
(2003).
Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites.
J Biol Inorg Chem,
8,
452-458.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
08-Apr-02
|
Release date:
|
20-May-03
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P32396
(HEMH_BACSU) -
Coproporphyrin III ferrochelatase from Bacillus subtilis (strain 168)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
310 a.a.
309 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.99.1.9
- coproporphyrin ferrochelatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Fe-coproporphyrin III + 2 H+ = coproporphyrin III + Fe2+
|
 |
 |
 |
 |
 |
Fe-coproporphyrin III
|
+
|
2
×
H(+)
|
=
|
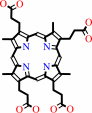 coproporphyrin III
coproporphyrin III
|
+
|
Fe(2+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Inorg Chem
8:452-458
(2003)
|
|
PubMed id:
|
|
|
|

|
| |
|
Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites.
|
|
D.Lecerof,
M.N.Fodje,
R.Alvarez León,
U.Olsson,
A.Hansson,
E.Sigfridsson,
U.Ryde,
M.Hansson,
S.Al-Karadaghi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ferrochelatase, the terminal enzyme in heme biosynthesis, catalyses metal
insertion into protoporphyrin IX. The location of the metal binding site with
respect to the bound porphyrin substrate and the mode of metal binding are of
central importance for understanding the mechanism of porphyrin metallation. In
this work we demonstrate that Zn(2+), which is commonly used as substrate in
assays of the ferrochelatase reaction, and Cd(2+), an inhibitor of the enzyme,
bind to the invariant amino acids His183 and Glu264 and water molecules, all
located within the porphyrin binding cleft. On the other hand, Mg(2+), which has
been shown to bind close to the surface at 7 A from His183, was largely absent
from its site. Activity measurements demonstrate that Mg(2+) has a stimulatory
effect on the enzyme, lowering K(M) for Zn(2+) from 55 to 24 micro M. Changing
one of the Mg(2+) binding residues, Glu272, to serine abolishes the effect of
Mg(2+). It is proposed that prior to metal insertion the metal may form a
sitting-atop (SAT) complex with the invariant His-Glu couple and the porphyrin.
Metal binding to the Mg(2+) site may stimulate metal release from the protein
ligands and its insertion into the porphyrin.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.D.Hansson,
T.Karlberg,
C.A.Söderberg,
S.Rajan,
M.J.Warren,
S.Al-Karadaghi,
S.E.Rigby,
and
M.Hansson
(2011).
Bacterial ferrochelatase turns human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase.
|
| |
J Biol Inorg Chem,
16,
235-242.
|
 |
|
|
|
|
 |
R.E.Davidson,
C.J.Chesters,
and
J.D.Reid
(2009).
Metal ion selectivity and substrate inhibition in the metal ion chelation catalyzed by human ferrochelatase.
|
| |
J Biol Chem,
284,
33795-33799.
|
 |
|
|
|
|
 |
G.A.Hunter,
M.P.Sampson,
and
G.C.Ferreira
(2008).
Metal ion substrate inhibition of ferrochelatase.
|
| |
J Biol Chem,
283,
23685-23691.
|
 |
|
|
|
|
 |
T.Karlberg,
M.D.Hansson,
R.K.Yengo,
R.Johansson,
H.O.Thorvaldsen,
G.C.Ferreira,
M.Hansson,
and
S.Al-Karadaghi
(2008).
Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues.
|
| |
J Mol Biol,
378,
1074-1083.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.E.Medlock,
T.A.Dailey,
T.A.Ross,
H.A.Dailey,
and
W.N.Lanzilotta
(2007).
A pi-helix switch selective for porphyrin deprotonation and product release in human ferrochelatase.
|
| |
J Mol Biol,
373,
1006-1016.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.A.Dailey,
C.K.Wu,
P.Horanyi,
A.E.Medlock,
W.Najahi-Missaoui,
A.E.Burden,
T.A.Dailey,
and
J.Rose
(2007).
Altered orientation of active site residues in variants of human ferrochelatase. Evidence for a hydrogen bond network involved in catalysis.
|
| |
Biochemistry,
46,
7973-7979.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.Yin,
L.X.Xu,
M.M.Cherney,
E.Raux-Deery,
A.A.Bindley,
A.Savchenko,
J.R.Walker,
M.E.Cuff,
M.J.Warren,
and
M.N.James
(2006).
Crystal structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus.
|
| |
J Struct Funct Genomics,
7,
37-50.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.D.Hansson,
M.Lindstam,
and
M.Hansson
(2006).
Crosstalk between metal ions in Bacillus subtilis ferrochelatase.
|
| |
J Biol Inorg Chem,
11,
325-333.
|
 |
|
|
|
|
 |
R.J.Sawers,
J.Viney,
P.R.Farmer,
R.R.Bussey,
G.Olsefski,
K.Anufrikova,
C.N.Hunter,
and
T.P.Brutnell
(2006).
The maize Oil yellow1 (Oy1) gene encodes the I subunit of magnesium chelatase.
|
| |
Plant Mol Biol,
60,
95.
|
 |
|
|
|
|
 |
S.Al-Karadaghi,
R.Franco,
M.Hansson,
J.A.Shelnutt,
G.Isaya,
and
G.C.Ferreira
(2006).
Chelatases: distort to select?
|
| |
Trends Biochem Sci,
31,
135-142.
|
 |
|
|
|
|
 |
Y.Shen,
and
U.Ryde
(2005).
Reaction mechanism of porphyrin metallation studied by theoretical methods.
|
| |
Chemistry,
11,
1549-1564.
|
 |
|
|
|
|
 |
Z.Shi,
and
G.C.Ferreira
(2004).
Probing the active site loop motif of murine ferrochelatase by random mutagenesis.
|
| |
J Biol Chem,
279,
19977-19986.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

