 |
PDBsum entry 1lbl

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.48
- indole-3-glycerol-phosphate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate + H+ = (1S,2R)- 1-C-(indol-3-yl)glycerol 3-phosphate + CO2 + H2O
|
 |
 |
 |
 |
 |
1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
|
=
|
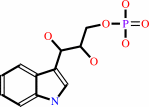 (1S,2R)- 1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)- 1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
 CO2
CO2
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
319:757-766
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
The catalytic mechanism of indole-3-glycerol phosphate synthase: crystal structures of complexes of the enzyme from Sulfolobus solfataricus with substrate analogue, substrate, and product.
|
|
M.Hennig,
B.D.Darimont,
J.N.Jansonius,
K.Kirschner.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Indoleglycerol phosphate synthase catalyzes the ring closure of an N-alkylated
anthranilate to a 3-alkyl indole derivative, a reaction requiring Lewis acid
catalysis in vitro. Here, we investigated the enzymatic reaction mechanism
through X-ray crystallography of complexes of the hyperthermostable enzyme from
Sulfolobus solfataricus with the substrate 1-(o-carboxyphenylamino)
1-deoxyribulose 5-phosphate, a substrate analogue and the product
indole-3-glycerol phosphate. The substrate and the substrate analogue are bound
to the active site in a similar, extended conformation between the previously
identified phosphate binding site and a hydrophobic pocket for the anthranilate
moiety. This binding mode is unproductive, because the carbon atoms that are to
be joined are too far apart. The indole ring of the bound product resides in a
second hydrophobic pocket adjacent to that of the anthranilate moiety of the
substrate. Although the hydrophobic moiety of the substrate moves during
catalysis from one hydrophobic pocket to the other, the triosephosphate moiety
remains rigidly bound to the same set of hydrogen-bonding residues.
Simultaneously, the catalytically important residues Lys53, Lys110 and Glu159
maintain favourable distances to the atoms of the ligand undergoing covalent
changes. On the basis of these data, the structures of two putative catalytic
intermediates were modelled into the active site. This new structural
information and the modelling studies provide further insight into the mechanism
of enzyme-catalyzed indole synthesis. The charged epsilon-amino group of Lys110
is the general acid, and the carboxylate group of Glu159 is the general base.
Lys53 guides the substrate undergoing conformational transitions during
catalysis, by forming a salt-bridge to the carboxylate group of its anthranilate
moiety.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 4.
Figure 4. Stereoview of the difference electron density of
the ligands bound to sIGPS, as it appears in electron density
omit maps. These were computed with phases resulting from
refinement, in which the ligand was omitted. The ligands are
shown in a ball-and-stick representation with carbon, oxygen,
nitrogen and phosphorus coloured white, red, blue and magenta,
respectively, (a) rCdRP on a 2.05 Å map, contoured at 4s.
(b) IGP on a 2.0 Å map, contoured at 4s. (c) CdRP and IGP
on a 2.4 Å map, contoured at 2.5s. The Figure was produced
with MOLOC.[29.]
|
 |
Figure 6.
Figure 6. The proposed roles of K53, K110 and E159 of sIGPS
in catalyzing the conversion of CdRP to IGP. Hydrogen bond
distances are given in Table 3. (*) Asymmetric carbon atoms
generated transiently. The arrows show how the reactions are
assisted by the indicated catalytic residues. See the text for
details. (a) The substrate bound unproductively as observed in
the sIGPS:CdRP complex ( Figure 3(c)), with C1-C2' DISTANCE=4.8
Å. (b) The intermediate I1 as in Figure 5(a). (c) The
intermediate I2 as in Figure 5(b). (d) The sIGPS:IGP complex as
in Figure 3(b).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2002,
319,
757-766)
copyright 2002.
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Shen,
F.Wang,
Y.Zhang,
Q.Huang,
S.Xu,
H.Hu,
J.Yue,
and
H.Wang
(2009).
A novel inhibitor of indole-3-glycerol phosphate synthase with activity against multidrug-resistant Mycobacterium tuberculosis.
|
| |
FEBS J,
276,
144-154.
|
 |
|
|
|
|
 |
J.Claren,
C.Malisi,
B.Höcker,
and
R.Sterner
(2009).
Establishing wild-type levels of catalytic activity on natural and artificial (beta alpha)8-barrel protein scaffolds.
|
| |
Proc Natl Acad Sci U S A,
106,
3704-3709.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.N.Alexandrova,
D.Röthlisberger,
D.Baker,
and
W.L.Jorgensen
(2008).
Catalytic mechanism and performance of computationally designed enzymes for Kemp elimination.
|
| |
J Am Chem Soc,
130,
15907-15915.
|
 |
|
|
|
|
 |
R.Das,
and
D.Baker
(2008).
Macromolecular modeling with rosetta.
|
| |
Annu Rev Biochem,
77,
363-382.
|
 |
|
|
|
|
 |
Z.Gu,
J.A.Zitzewitz,
and
C.R.Matthews
(2007).
Mapping the structure of folding cores in TIM barrel proteins by hydrogen exchange mass spectrometry: the roles of motif and sequence for the indole-3-glycerol phosphate synthase from Sulfolobus solfataricus.
|
| |
J Mol Biol,
368,
582-594.
|
 |
|
|
|
|
 |
A.Gutteridge,
and
J.M.Thornton
(2005).
Understanding nature's catalytic toolkit.
|
| |
Trends Biochem Sci,
30,
622-629.
|
 |
|
|
|
|
 |
D.Mazumder-Shivakumar,
and
T.C.Bruice
(2004).
Molecular dynamics studies of ground state and intermediate of the hyperthermophilic indole-3-glycerol phosphate synthase.
|
| |
Proc Natl Acad Sci U S A,
101,
14379-14384.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

