 |
PDBsum entry 1l4h

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.21
- nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Corrin Biosynthesis (part 8)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5,6-dimethylbenzimidazole + nicotinate beta-D-ribonucleotide = alpha- ribazole 5'-phosphate + nicotinate + H+
|
 |
 |
 |
 |
 |
5,6-dimethylbenzimidazole
Bound ligand (Het Group name = )
matches with 81.82% similarity
|
+
|
nicotinate beta-D-ribonucleotide
|
=
|
alpha- ribazole 5'-phosphate
Bound ligand (Het Group name = )
matches with 76.92% similarity
|
+
|
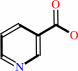 nicotinate
nicotinate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biol Chem
277:41120-41127
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
|
|
C.G.Cheong,
J.C.Escalante-Semerena,
I.Rayment.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Nicotinate mononucleotide (NaMN):5,6-dimethylbenzimidazole (DMB)
phosphoribosyltransferase (CobT) from Salmonella enterica plays a central role
in the synthesis of alpha-ribazole-5'-phosphate, an intermediate for the lower
ligand of cobalamin. In earlier studies it proved difficult to obtain the
structure of CobT bound to NaMN because it is hydrolyzed in the crystal lattice
in the absence of the second substrate DMB. In an effort to map the reaction
pathway of this enzyme, NaMN was captured in the active site with the substrate
analogs 4,5-dimethyl-1,2-phenylenediamine, 4-methylcatechol, indole,
3,4-dimethylaniline, 2,5-dimethylaniline, 3,4-dimethylphenol, and
2-amino-p-cresol. Structures of these complexes reveal that they exclude water
molecules responsible for the hydrolysis from the active site. These structures,
together with the early complexes with alpha-ribazole-5'-phosphate and DMB,
provide a complete description of the reaction pathway. They demonstrate that
the nicotinate moiety and phosphate do not appear to move significantly between
reactants and products but that the aromatic base and ribose moiety each move
approximately 1.2 A toward each other in the transformation. This study also
reveals that, like many other nucleotide binding proteins, coordination of DMB
is accompanied by a disorder-order transition in a surface loop. The structure
of apo-CobT is also reported.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

