 |
PDBsum entry 1kuu

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics, unknown function
|
PDB id
|
|

|

|
1kuu
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.4.10
- Imp cyclohydrolase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Purine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + H2O = 5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide
|
 |
 |
 |
 |
 |
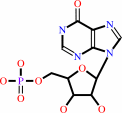 IMP
IMP
|
+
|
H2O
|
=
|
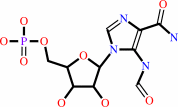 5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide
5-formamido-1-(5-phospho-D-ribosyl)imidazole-4-carboxamide
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
48:141-143
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Methanobacterium thermoautotrophicum conserved protein MTH1020 reveals an NTN-hydrolase fold.
|
|
V.Saridakis,
D.Christendat,
A.Thygesen,
C.H.Arrowsmith,
A.M.Edwards,
E.F.Pai.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1. A: Ribbon diagram of a subunit of MTH1020. The
protein is composed of a single domain with an  - -
  - -
 core.
The secondary structure elements are numbered. B: Structure
superposition of MTH1020 (green) with a member of the
NTN-hydrolase family (blue) showing the similarity of the
structure of MTH1020 with NTN-hydrolases. The arrows depict Arg5
and the N-terminal nucleophile of the NTN-hydrolase member in
ball and stick form showing the similarity in position of Arg5
with the N-terminal nucleophile. The programs MOLSCRIPT and
RASTER 3D were used in the production of the figures. core.
The secondary structure elements are numbered. B: Structure
superposition of MTH1020 (green) with a member of the
NTN-hydrolase family (blue) showing the similarity of the
structure of MTH1020 with NTN-hydrolases. The arrows depict Arg5
and the N-terminal nucleophile of the NTN-hydrolase member in
ball and stick form showing the similarity in position of Arg5
with the N-terminal nucleophile. The programs MOLSCRIPT and
RASTER 3D were used in the production of the figures.
|
 |
|
|

|
| |
The above figure is
reprinted
by permission from John Wiley & Sons, Inc.:
Proteins
(2002,
48,
141-143)
copyright 2002.
|
|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Zhang,
M.Morar,
and
S.E.Ealick
(2008).
Structural biology of the purine biosynthetic pathway.
|
| |
Cell Mol Life Sci,
65,
3699-3724.
|
 |
|
|
|
|
 |
Y.N.Kang,
A.Tran,
R.H.White,
and
S.E.Ealick
(2007).
A novel function for the N-terminal nucleophile hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase.
|
| |
Biochemistry,
46,
5050-5062.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

