 |
PDBsum entry 1kut

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Structural genomics, ligase
|
PDB id
|
|

|

|
1kut
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Structural genomics, ligase
|
 |
|
Title:
|
 |
Structural genomics, protein tm1243, (saicar synthetase)
|
|
Structure:
|
 |
Phosphoribosylaminoimidazole-succinocarboxamide synthase. Chain: a, b. Synonym: saicar synthetase. Engineered: yes
|
|
Source:
|
 |
Thermotoga maritima. Organism_taxid: 2336. Gene: tm1243. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Biol. unit:
|
 |
Dimer (from
)
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.245
|
R-free:
|
0.281
|
|
|
Authors:
|
 |
R.Zhang,T.Skarina,S.Beasley,A.Edwards,A.Joachimiak,A.Savchenko, Midwest Center For Structural Genomics (Mcsg)
|
Key ref:

|
 |
R.Zhang
et al.
(2006).
Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallograph Sect F Struct Biol Cryst Commun,
62,
335-339.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
22-Jan-02
|
Release date:
|
14-Aug-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.6.3.2.6
- phosphoribosylaminoimidazolesuccinocarboxamide synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Purine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate + L-aspartate + ATP = (2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamido]succinate + ADP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
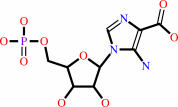 5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate
5-amino-1-(5-phospho-D-ribosyl)imidazole-4-carboxylate
|
+
|
 L-aspartate
L-aspartate
|
+
|
 ATP
ATP
|
=
|
(2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamido]succinate
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallograph Sect F Struct Biol Cryst Commun
62:335-339
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
|
|
R.Zhang,
T.Skarina,
E.Evdokimova,
A.Edwards,
A.Savchenko,
R.Laskowski,
M.E.Cuff,
A.Joachimiak.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
As a part of a structural genomics program, the 2.2 angstroms resolution crystal
structure of the PurC gene product from Thermotoga maritima has been solved.
This 26.2 kDa protein belongs to the
phophoribosylaminoimidazole-succinocarboxamide or SAICAR synthase family of
enzymes, the members of which are involved in de novo purine biosynthesis.
SAICAR synthase can be divided into three subdomains: two alpha+beta regions
exhibiting structural homology with ATP-binding proteins and a carboxy-terminal
subdomain of two alpha-helices. The asymmetric unit contains two copies of the
protein which are covalently linked by a disulfide bond between Cys126(A) and
Cys126(B). This 230-amino-acid protein exhibits high structural homology with
SAICAR synthase from baker's yeast. The protein structure is described and
compared with that of the ATP-SAICAR synthase complex from yeast.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1 Ribbon schematic of the T. maritima SAICAR synthase
monomer. The peptide chain is colored blue to red from the N- to
the C-terminus. Conserved residue Glu172 is labeled E172; Cys126
is labeled C126 and is cross-linked to its counterpart in the
second molecule of the asymmetric unit. All ribbon figures were
generated using PyMOL (DeLano, 2002[DeLano, W. L. (2002). The
PyMOL Molecular Graphics System. http://www.pymol.org .]).
|
 |
Figure 3.
Figure 3 Dimeric SAICAR synthase. The covalent dimer is
pictured here in stereo with one peptide chain in red and the
other in blue. The amino-termini are labeled N and the
carboxy-termini are labeled C. The disulfide bridge is located
across a (noncrystallographic) twofold axis and is shown as
yellow sticks.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by the IUCr:
Acta Crystallograph Sect F Struct Biol Cryst Commun
(2006,
62,
335-339)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.D.Ginder,
D.J.Binkowski,
H.J.Fromm,
and
R.B.Honzatko
(2006).
Nucleotide complexes of Escherichia coli phosphoribosylaminoimidazole succinocarboxamide synthetase.
|
| |
J Biol Chem,
281,
20680-20688.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |
|

