 |
PDBsum entry 1kog

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase/RNA
|
 |
|
Title:
|
 |
Crystal structure of e. Coli threonyl-tRNA synthetase interacting with the essential domain of its mRNA operator
|
|
Structure:
|
 |
Threonyl-tRNA synthetase mRNA. Chain: i, j, k, l, m, n, o, p. Engineered: yes. Mutation: yes. Other_details: domain d2 of the trs mRNA operator from e.Coli (nucleotides -49 to -13). Threonyl-tRNA synthetase. Chain: a, b, c, d, e, f, g, h. Fragment: catalytic and anticodon binding domains (residues 242 to
|
|
Source:
|
 |
Synthetic: yes. Other_details: this is the natural sequence of domain d2 of the trs mRNA operator from e. Coli, covering residues -49 to -13 (69 to 105 in the present coordinate file) of e.Coli trs mRNA, except for the first 3 base pairs. It was synthesized in vitro by t7 transcription.. Escherichia coli. Organism_taxid: 562. Expressed in: escherichia coli.
|
|
Biol. unit:
|
 |
 Tetramer (from PDB file)
Tetramer (from PDB file)
|
|
Resolution:
|
 |
|
3.50Å
|
R-factor:
|
0.251
|
R-free:
|
0.287
|
|
|
Authors:
|
 |
A.Torres-Larrios,A.C.Dock-Bregeon,P.Romby,B.Rees,R.Sankaranarayanan, J.Caillet,M.Springer,C.Ehresmann,B.Ehresmann,D.Moras
|
Key ref:

|
 |
A.Torres-Larios
et al.
(2002).
Structural basis of translational control by Escherichia coli threonyl tRNA synthetase.
Nat Struct Biol,
9,
343-347.
PubMed id:
|
 |
|
Date:
|
 |
|
20-Dec-01
|
Release date:
|
26-Apr-02
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0A8M3
(SYT_ECOLI) -
Threonine--tRNA ligase from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
642 a.a.
401 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|

|

|
|
|
|
G-G-C-G-U-A-U-G-U-G-A-U-C-U-U-U-C-G-U-G-U-G-G-G-U-C-A-C-C-A-C-U-G-C-G-C-C
37 bases
|
|
|
|
G-G-C-G-U-A-U-G-U-G-A-U-C-U-U-U-C-G-U-G-U-G-G-G-U-C-A-C-C-A-C-U-G-C-G-C-C
37 bases
|
|
|
|
G-G-C-G-U-A-U-G-U-G-A-U-C-U-U-U-C-G-U-G-U-G-G-G-U-C-A-C-C-A-C-U-G-C-G-C-C
37 bases
|
|
|
|
G-G-C-G-U-A-U-G-U-G-A-U-C-U-U-U-C-G-U-G-U-G-G-G-U-C-A-C-C-A-C-U-G-C-G-C-C
37 bases
|
|
|
|
G-G-C-G-U-A-U-G-U-G-A-U-C-U-U-U-C-G-U-G-U-G-G-G-U-C-A-C-C-A-C-U-G-C-G-C-C
37 bases
|
|
|
|
G-G-C-G-U-A-U-G-U-G-A-U-C-U-U-U-C-G-U-G-U-G-G-G-U-C-A-C-C-A-C-U-G-C-G-C-C
37 bases
|
|
|
|
G-G-C-G-U-A-U-G-U-G-A-U-C-U-U-U-C-G-U-G-U-G-G-G-U-C-A-C-C-A-C-U-G-C-G-C-C
37 bases
|
|
|
|
G-G-C-G-U-A-U-G-U-G-A-U-C-U-U-U-C-G-U-G-U-G-G-G-U-C-A-C-C-A-C-U-G-C-G-C-C
37 bases
|
|

|
 |
 |
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.3
- threonine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Thr) + L-threonine + ATP = L-threonyl-tRNA(Thr) + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
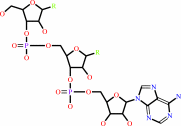 tRNA(Thr)
tRNA(Thr)
|
+
|
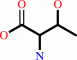 L-threonine
L-threonine
|
+
|
 ATP
ATP
|
=
|
L-threonyl-tRNA(Thr)
Bound ligand (Het Group name = )
matches with 55.88% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Struct Biol
9:343-347
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of translational control by Escherichia coli threonyl tRNA synthetase.
|
|
A.Torres-Larios,
A.C.Dock-Bregeon,
P.Romby,
B.Rees,
R.Sankaranarayanan,
J.Caillet,
M.Springer,
C.Ehresmann,
B.Ehresmann,
D.Moras.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Escherichia coli threonyl-tRNA synthetase (ThrRS) represses the translation of
its own messenger RNA by binding to an operator located upstream of the
initiation codon. The crystal structure of the complex between the core of ThrRS
and the essential domain of the operator shows that the mRNA uses the
recognition mode of the tRNA anticodon loop to initiate binding. The final
positioning of the operator, upon which the control mechanism is based, relies
on a characteristic RNA motif adapted to the enzyme surface. The finding of
other thrS operators that have this conserved motif leads to a generalization of
this regulatory mechanism to a subset of Gram-negative bacteria.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Zhong,
H.Tang,
and
S.Zhang
(2010).
RNAMotifScan: automatic identification of RNA structural motifs using secondary structural alignment.
|
| |
Nucleic Acids Res,
38,
e176.
|
 |
|
|
|
|
 |
P.Babitzke,
C.S.Baker,
and
T.Romeo
(2009).
Regulation of translation initiation by RNA binding proteins.
|
| |
Annu Rev Microbiol,
63,
27-44.
|
 |
|
|
|
|
 |
C.D.Hausmann,
and
M.Ibba
(2008).
Aminoacyl-tRNA synthetase complexes: molecular multitasking revealed.
|
| |
FEMS Microbiol Rev,
32,
705-721.
|
 |
|
|
|
|
 |
E.E.Regulski,
R.H.Moy,
Z.Weinberg,
J.E.Barrick,
Z.Yao,
W.L.Ruzzo,
and
R.R.Breaker
(2008).
A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism.
|
| |
Mol Microbiol,
68,
918-932.
|
 |
|
|
|
|
 |
R.Arreola,
A.Vega-Miranda,
A.Gómez-Puyou,
R.Pérez-Montfort,
E.Merino-Pérez,
and
A.Torres-Larios
(2008).
Expression, purification and preliminary X-ray diffraction studies of the transcriptional factor PyrR from Bacillus halodurans.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
692-696.
|
 |
|
|
|
|
 |
S.Shimizu,
E.C.Juan,
Y.I.Miyashita,
Y.Sato,
M.M.Hoque,
K.Suzuki,
M.Yogiashi,
M.Tsunoda,
A.C.Dock-Bregeon,
D.Moras,
T.Sekiguchi,
and
A.Takénaka
(2008).
Crystallization and preliminary crystallographic studies of putative threonyl-tRNA synthetases from Aeropyrum pernix and Sulfolobus tokodaii.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
903-910.
|
 |
|
|
|
|
 |
J.Caillet,
M.Graffe,
F.Eyermann,
P.Romby,
and
M.Springer
(2007).
Mutations in residues involved in zinc binding in the catalytic site of Escherichia coli threonyl-tRNA synthetase confer a dominant lethal phenotype.
|
| |
J Bacteriol,
189,
6839-6848.
|
 |
|
|
|
|
 |
R.T.Batey
(2006).
Structures of regulatory elements in mRNAs.
|
| |
Curr Opin Struct Biol,
16,
299-306.
|
 |
|
|
|
|
 |
X.L.Yang,
F.J.Otero,
K.L.Ewalt,
J.Liu,
M.A.Swairjo,
C.Köhrer,
U.L.RajBhandary,
R.J.Skene,
D.E.McRee,
and
P.Schimmel
(2006).
Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis.
|
| |
EMBO J,
25,
2919-2929.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Lescoute,
N.B.Leontis,
C.Massire,
and
E.Westhof
(2005).
Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments.
|
| |
Nucleic Acids Res,
33,
2395-2409.
|
 |
|
|
|
|
 |
L.Jenner,
P.Romby,
B.Rees,
C.Schulze-Briese,
M.Springer,
C.Ehresmann,
B.Ehresmann,
D.Moras,
G.Yusupova,
and
M.Yusupov
(2005).
Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding.
|
| |
Science,
308,
120-123.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Beebe,
E.Merriman,
L.Ribas De Pouplana,
and
P.Schimmel
(2004).
A domain for editing by an archaebacterial tRNA synthetase.
|
| |
Proc Natl Acad Sci U S A,
101,
5958-5963.
|
 |
|
|
|
|
 |
N.Kim,
E.A.Marcus,
Y.Wen,
D.L.Weeks,
D.R.Scott,
H.C.Jung,
I.S.Song,
and
G.Sachs
(2004).
Genes of Helicobacter pylori regulated by attachment to AGS cells.
|
| |
Infect Immun,
72,
2358-2368.
|
 |
|
|
|
|
 |
A.R.Ferré-D'Amaré
(2003).
RNA-modifying enzymes.
|
| |
Curr Opin Struct Biol,
13,
49-55.
|
 |
|
|
|
|
 |
A.Serganov,
A.Polonskaia,
B.Ehresmann,
C.Ehresmann,
and
D.J.Patel
(2003).
Ribosomal protein S15 represses its own translation via adaptation of an rRNA-like fold within its mRNA.
|
| |
EMBO J,
22,
1898-1908.
|
 |
|
|
|
|
 |
J.Caillet,
T.Nogueira,
B.Masquida,
F.Winter,
M.Graffe,
A.C.Dock-Brégeon,
A.Torres-Larios,
R.Sankaranarayanan,
E.Westhof,
B.Ehresmann,
C.Ehresmann,
P.Romby,
and
M.Springer
(2003).
The modular structure of Escherichia coli threonyl-tRNA synthetase as both an enzyme and a regulator of gene expression.
|
| |
Mol Microbiol,
47,
961-974.
|
 |
|
|
|
|
 |
N.B.Leontis,
and
E.Westhof
(2003).
Analysis of RNA motifs.
|
| |
Curr Opin Struct Biol,
13,
300-308.
|
 |
|
|
|
|
 |
P.J.Schlax,
and
D.J.Worhunsky
(2003).
Translational repression mechanisms in prokaryotes.
|
| |
Mol Microbiol,
48,
1157-1169.
|
 |
|
|
|
|
 |
P.Romby,
and
M.Springer
(2003).
Bacterial translational control at atomic resolution.
|
| |
Trends Genet,
19,
155-161.
|
 |
|
|
|
|
 |
U.Stelzl,
J.M.Zengel,
M.Tovbina,
M.Walker,
K.H.Nierhaus,
L.Lindahl,
and
D.J.Patel
(2003).
RNA-structural mimicry in Escherichia coli ribosomal protein L4-dependent regulation of the S10 operon.
|
| |
J Biol Chem,
278,
28237-28245.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

