 |
PDBsum entry 1kgo

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Metal binding protein
|
PDB id
|
|

|

|
1kgo
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Metal binding protein
|
 |
|
Title:
|
 |
R2f from corynebacterium ammoniagenes in its reduced, fe containing, form
|
|
Structure:
|
 |
Ribonucleotide reductase protein r2f. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Corynebacterium ammoniagenes. Organism_taxid: 1697. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Biol. unit:
|
 |
 Dimer (from
)
Dimer (from
)
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.163
|
R-free:
|
0.238
|
|
|
Authors:
|
 |
M.Hogbom,Y.Huque,B.M.Sjoberg,P.Nordlund
|
Key ref:

|
 |
M.Högbom
et al.
(2002).
Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
Biochemistry,
41,
1381-1389.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
28-Nov-01
|
Release date:
|
21-Dec-01
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O69274
(O69274_CORAM) -
Ribonucleoside-diphosphate reductase subunit beta from Corynebacterium ammoniagenes
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
329 a.a.
296 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.17.4.1
- ribonucleoside-diphosphate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside 5'-diphosphate + [thioredoxin]-disulfide + H2O = a ribonucleoside 5'-diphosphate + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
 2'-deoxyribonucleoside diphosphate
2'-deoxyribonucleoside diphosphate
|
+
|
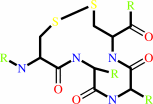 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
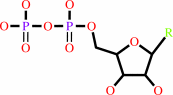 ribonucleoside diphosphate
ribonucleoside diphosphate
|
+
|
 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(3+) or adenosylcob(III)alamin or Mn(2+)
|
 |
 |
 |
 |
 |
Fe(3+)
|
or
|
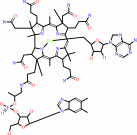 adenosylcob(III)alamin
adenosylcob(III)alamin
|
or
|
Mn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
41:1381-1389
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the di-iron/radical protein of ribonucleotide reductase from Corynebacterium ammoniagenes.
|
|
M.Högbom,
Y.Huque,
B.M.Sjöberg,
P.Nordlund.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Ribonucleotide reductase (RNR) is the enzyme performing de novo production of
the four deoxyribonucleotides needed for DNA synthesis. All mammals as well as
some prokaryotes express the class I enzyme which is an alpha(2)beta(2) protein.
The smaller of the homodimers, denoted R2, contains a di-iron carboxylate site
which, upon reaction with molecular oxygen, generates a stable tyrosyl radical
needed for catalysis. The three-dimensional structure of the oxidized class Ib
RNR R2 from Corynebacterium ammoniagenes has been determined at 1.85 A
resolution and refined to an R-value of 15.8% (R(free) = 21.3%). In addition,
structures of both the reduced iron-containing, and manganese-substituted
protein have been solved. The C. ammoniagenes R2 has been proposed to be
manganese-dependent. The present structure provides evidence that manganese is
not oxidized by the protein, in agreement with recent biochemical data, and that
no obvious structural abnormalities are seen in the oxidized and reduced
iron-containing forms, giving further support that the protein is indeed an
iron-dependent RNR R2. The di-manganese structure also provides an explanation
for the magnetic properties of this site. The structure of the oxidized C.
ammoniagenes R2 also reveals an additional water molecule bridging the radical
and the iron site, which has not previously been seen in any other R2 structure
and which might have important mechanistic implications.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.Högbom
(2011).
Metal use in ribonucleotide reductase R2, di-iron, di-manganese and heterodinuclear--an intricate bioinorganic workaround to use different metals for the same reaction.
|
| |
Metallomics,
3,
110-120.
|
 |
|
|
|
|
 |
A.K.Boal,
J.A.Cotruvo,
J.Stubbe,
and
A.C.Rosenzweig
(2010).
Structural basis for activation of class Ib ribonucleotide reductase.
|
| |
Science,
329,
1526-1530.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
B.M.Sjöberg
(2010).
Biochemistry. A never-ending story.
|
| |
Science,
329,
1475-1476.
|
 |
|
|
|
|
 |
P.Stolle,
O.Barckhausen,
W.Oehlmann,
N.Knobbe,
C.Vogt,
A.J.Pierik,
N.Cox,
P.P.Schmidt,
E.J.Reijerse,
W.Lubitz,
and
G.Auling
(2010).
Homologous expression of the nrdF gene of Corynebacterium ammoniagenes strain ATCC 6872 generates a manganese-metallocofactor (R2F) and a stable tyrosyl radical (Y˙) involved in ribonucleotide reduction.
|
| |
FEBS J,
277,
4849-4862.
|
 |
|
|
|
|
 |
N.Voevodskaya,
F.Lendzian,
O.Sanganas,
A.Grundmeier,
A.Gräslund,
and
M.Haumann
(2009).
Redox Intermediates of the Mn-Fe Site in Subunit R2 of Chlamydia trachomatis Ribonucleotide Reductase: AN X-RAY ABSORPTION AND EPR STUDY.
|
| |
J Biol Chem,
284,
4555-4566.
|
 |
|
|
|
|
 |
L.L.Kelley,
B.D.Dillard,
W.Tempel,
L.Chen,
N.Shaw,
D.Lee,
M.G.Newton,
F.J.Sugar,
F.E.Jenney,
H.S.Lee,
C.Shah,
F.L.Poole,
M.W.Adams,
J.S.Richardson,
D.C.Richardson,
Z.J.Liu,
B.C.Wang,
and
J.Rose
(2007).
Structure of the hypothetical protein PF0899 from Pyrococcus furiosus at 1.85 A resolution.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
549-552.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Galander,
M.Uppsten,
U.Uhlin,
and
F.Lendzian
(2006).
Orientation of the tyrosyl radical in Salmonella typhimurium class Ib ribonucleotide reductase determined by high field EPR of R2F single crystals.
|
| |
J Biol Chem,
281,
31743-31752.
|
 |
|
|
|
|
 |
P.Nordlund,
and
P.Reichard
(2006).
Ribonucleotide reductases.
|
| |
Annu Rev Biochem,
75,
681-706.
|
 |
|
|
|
|
 |
E.Torrents,
M.Sahlin,
D.Biglino,
A.Gräslund,
and
B.M.Sjöberg
(2005).
Efficient growth inhibition of Bacillus anthracis by knocking out the ribonucleotide reductase tyrosyl radical.
|
| |
Proc Natl Acad Sci U S A,
102,
17946-17951.
|
 |
|
|
|
|
 |
K.R.Strand,
S.Karlsen,
M.Kolberg,
A.K.Røhr,
C.H.Görbitz,
and
K.K.Andersson
(2004).
Crystal structural studies of changes in the native dinuclear iron center of ribonucleotide reductase protein R2 from mouse.
|
| |
J Biol Chem,
279,
46794-46801.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Stenmark,
M.Högbom,
C.Roshick,
G.McClarty,
and
P.Nordlund
(2004).
Crystals of the ribonucleotide reductase R2 protein from Chlamydia trachomatis obtained by heavy-atom co-crystallization.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
376-378.
|
 |
|
|
|
|
 |
X.Liu,
and
E.C.Theil
(2004).
Ferritin reactions: direct identification of the site for the diferric peroxide reaction intermediate.
|
| |
Proc Natl Acad Sci U S A,
101,
8557-8562.
|
 |
|
|
|
|
 |
B.S.Cooperman
(2003).
Oligopeptide inhibition of class I ribonucleotide reductases.
|
| |
Biopolymers,
71,
117-131.
|
 |
|
|
|
|
 |
D.G.Kehres,
and
M.E.Maguire
(2003).
Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria.
|
| |
FEMS Microbiol Rev,
27,
263-290.
|
 |
|
|
|
|
 |
K.R.Strand,
S.Karlsen,
and
K.K.Andersson
(2002).
Cobalt substitution of mouse R2 ribonucleotide reductase as a model for the reactive diferrous state Spectroscopic and structural evidence for a ferromagnetically coupled dinuclear cobalt cluster.
|
| |
J Biol Chem,
277,
34229-34238.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

