 |
PDBsum entry 1k7f

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.4.2.1.20
- tryptophan synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate + L-serine = D-glyceraldehyde 3-phosphate + L-tryptophan + H2O
|
 |
 |
 |
 |
 |
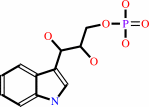 (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
 L-serine
L-serine
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
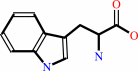 L-tryptophan
L-tryptophan
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 52.17% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
277:10647-10652
(2002)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
|
|
M.Weyand,
I.Schlichting,
A.Marabotti,
A.Mozzarelli.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Tryptophan synthase is a bifunctional alpha(2)beta(2) complex catalyzing the
last two steps of l-tryptophan biosynthesis. The natural substrates of the
alpha-subunit indole- 3-glycerolphosphate and glyceraldehyde-3-phosphate, and
the substrate analogs indole-3-propanolphosphate and
dl-alpha-glycerol-3-phosphate are allosteric effectors of the beta-subunit
activity. It has been shown recently, that the indole-3-acetyl amino acids
indole-3-acetylglycine and indole-3-acetyl-l-aspartic acid are both
alpha-subunit inhibitors and beta-subunit allosteric effectors, whereas
indole-3-acetyl-l-valine is only an alpha-subunit inhibitor (Marabotti, A.,
Cozzini, P., and Mozzarelli, A. (2000) Biochim. Biophys. Acta 1476, 287-299).
The crystal structures of tryptophan synthase complexed with
indole-3-acetylglycine and indole-3-acetyl-l-aspartic acid show that both
ligands bind to the active site such that the carboxylate moiety is positioned
similarly as the phosphate group of the natural substrates. As a consequence,
the residues of the alpha-active site that interact with the ligands are the
same as observed in the indole 3-glycerolphosphate-enzyme complex. Ligand
binding leads to closure of loop alphaL6 of the alpha-subunit, a key structural
element of intersubunit communication. This is in keeping with the allosteric
role played by these compounds. The structure of the enzyme complex with
indole-3-acetyl-l-valine is quite different. Due to the hydrophobic lateral
chain, this molecule adopts a new orientation in the alpha-active site. In this
case, closure of loop alphaL6 is no longer observed, in agreement with its
functioning only as an inhibitor of the alpha-subunit reaction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. 2mF[o]  DF[c]
electron density for the TRPSIAAA structures at the DF[c]
electron density for the TRPSIAAA structures at the  -active
site. The density is shown at 1 -active
site. The density is shown at 1  -contouring
for the IAAA molecule, -contouring
for the IAAA molecule,  Glu49, Glu49,  Asp60, Asp60,  Thr183, and
for water molecules, which form hydrogen bonds with the Thr183, and
for water molecules, which form hydrogen bonds with the  -ligand.
The hydrogen bonds of the enzymatic important amino acids -ligand.
The hydrogen bonds of the enzymatic important amino acids  Glu49 and Glu49 and
 Asp60 are
shown as dashed lines (see Table II). The figure was prepared
using "BOBSCRIPT" (40), "MOLSCRIPT" (41), and "RASTER3D" (42,
43). A, TRPSIAD structure. B, TRPSIAG structure. The second
conformation of Asp60 are
shown as dashed lines (see Table II). The figure was prepared
using "BOBSCRIPT" (40), "MOLSCRIPT" (41), and "RASTER3D" (42,
43). A, TRPSIAD structure. B, TRPSIAG structure. The second
conformation of  Glu49 is
shown in orange. C, TRPSIAV structure. Glu49 is
shown in orange. C, TRPSIAV structure.
|
 |
Figure 2.
Fig. 2. Stereo plot of the structure superposition of
TRPSIPP , TRPSIAD, and TRPSIAV. The C[  ]-atom
trace is shown for TRPSIPP; the ]-atom
trace is shown for TRPSIPP; the  -subunit is
colored in gray, loops -subunit is
colored in gray, loops  L2 and L2 and  L6 in cyan,
and the L6 in cyan,
and the  -subunit in
pink. The IPP, IAD, and IAV ligand C-atoms are colored in
yellow, green, or orange, respectively. Nitrogen atoms are
colored in blue, oxygen atoms are colored in red, and phosphate
atoms are colored in magenta. The figure was prepared using
MOLSCRIPT (41) and RASTER3D (42). -subunit in
pink. The IPP, IAD, and IAV ligand C-atoms are colored in
yellow, green, or orange, respectively. Nitrogen atoms are
colored in blue, oxygen atoms are colored in red, and phosphate
atoms are colored in magenta. The figure was prepared using
MOLSCRIPT (41) and RASTER3D (42).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2002,
277,
10647-10652)
copyright 2002.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Raboni,
S.Bettati,
and
A.Mozzarelli
(2009).
Tryptophan synthase: a mine for enzymologists.
|
| |
Cell Mol Life Sci,
66,
2391-2403.
|
 |
|
|
|
|
 |
M.F.Dunn,
D.Niks,
H.Ngo,
T.R.Barends,
and
I.Schlichting
(2008).
Tryptophan synthase: the workings of a channeling nanomachine.
|
| |
Trends Biochem Sci,
33,
254-264.
|
 |
|
|
|
|
 |
Y.Hioki,
K.Ogasahara,
S.J.Lee,
J.Ma,
M.Ishida,
Y.Yamagata,
Y.Matsuura,
M.Ota,
M.Ikeguchi,
S.Kuramitsu,
and
K.Yutani
(2004).
The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors.
|
| |
Eur J Biochem,
271,
2624-2635.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |
|

