 |
PDBsum entry 1jqc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
1jqc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.17.4.1
- ribonucleoside-diphosphate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2'-deoxyribonucleoside 5'-diphosphate + [thioredoxin]-disulfide + H2O = a ribonucleoside 5'-diphosphate + [thioredoxin]-dithiol
|
 |
 |
 |
 |
 |
 2'-deoxyribonucleoside diphosphate
2'-deoxyribonucleoside diphosphate
|
+
|
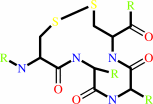 thioredoxin disulfide
thioredoxin disulfide
|
+
|
H(2)O
|
=
|
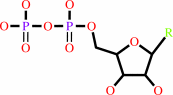 ribonucleoside diphosphate
ribonucleoside diphosphate
|
+
|
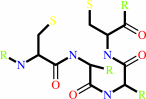 thioredoxin
thioredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(3+) or adenosylcob(III)alamin or Mn(2+)
|
 |
 |
 |
 |
 |
Fe(3+)
|
or
|
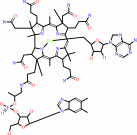 adenosylcob(III)alamin
adenosylcob(III)alamin
|
or
|
Mn(2+)
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biol Inorg Chem
6:315-323
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of oxidized dinuclear manganese centres in Mn-substituted class I ribonucleotide reductase from Escherichia coli: carboxylate shifts with implications for O2 activation and radical generation.
|
|
M.Högbom,
M.E.Andersson,
P.Nordlund.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The di-iron carboxylate proteins constitute a diverse class of non-heme iron
enzymes performing a multitude of redox reactions. These reactions usually
involve high-valent Fe-oxo species and are thought to be controlled by
carboxylate shifts. Owing to their short lifetime, the intermediate structures
have so far escaped structural characterization by X-ray crystallography. In an
attempt to map the carboxylate conformations available to the protein during
different redox states and different ligand environments, we have studied
metal-substituted forms of the R2 protein of ribonucleotide reductase from
Escherichia coli. In the present work we have solved the crystal structures of
Mn-substituted R2 oxidized in two different ways. Oxidation was performed using
either nitric oxide or a combination of hydrogen peroxide and hydroxylamine. The
two structures are virtually identical, indicating that the oxidation states are
the same, most likely a mixed-valent MnII-MnIII centre. One of the carboxylate
ligands (D84) adopts a new, so far unseen, conformation, which could participate
in the mechanism for radical generation in R2. E238 adopts a bridging-chelating
conformation proposed to be important for proper O2 activation but not
previously observed in the wild-type enzyme. Probable catalase activity was also
observed during the oxidation with H2O2, indicating mechanistic similarities to
the di-Mn catalases.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.E.Martin,
and
J.A.Imlay
(2011).
The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation.
|
| |
Mol Microbiol,
80,
319-334.
|
 |
|
|
|
|
 |
M.Högbom
(2011).
Metal use in ribonucleotide reductase R2, di-iron, di-manganese and heterodinuclear--an intricate bioinorganic workaround to use different metals for the same reaction.
|
| |
Metallomics,
3,
110-120.
|
 |
|
|
|
|
 |
C.L.Berthold,
H.Wang,
S.Nordlund,
and
M.Högbom
(2009).
Mechanism of ADP-ribosylation removal revealed by the structure and ligand complexes of the dimanganese mono-ADP-ribosylhydrolase DraG.
|
| |
Proc Natl Acad Sci U S A,
106,
14247-14252.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.S.Andersson,
and
M.Högbom
(2009).
A Mycobacterium tuberculosis ligand-binding Mn/Fe protein reveals a new cofactor in a remodeled R2-protein scaffold.
|
| |
Proc Natl Acad Sci U S A,
106,
5633-5638.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.S.Covarrubias,
T.Bergfors,
T.A.Jones,
and
M.Högbom
(2006).
Structural mechanics of the pH-dependent activity of beta-carbonic anhydrase from Mycobacterium tuberculosis.
|
| |
J Biol Chem,
281,
4993-4999.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Hoffmann,
K.Eitner,
M.von Grotthuss,
L.Rychlewski,
E.Banachowicz,
T.Grabarkiewicz,
T.Szkoda,
and
A.Kolinski
(2006).
Three dimensional model of severe acute respiratory syndrome coronavirus helicase ATPase catalytic domain and molecular design of severe acute respiratory syndrome coronavirus helicase inhibitors.
|
| |
J Comput Aided Mol Des,
20,
305-319.
|
 |
|
|
|
|
 |
M.Sommerhalter,
L.Saleh,
J.M.Bollinger,
and
A.C.Rosenzweig
(2005).
Structure of Escherichia coli ribonucleotide reductase R2 in space group P6122.
|
| |
Acta Crystallogr D Biol Crystallogr,
61,
1649-1654.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.Högbom,
M.Galander,
M.Andersson,
M.Kolberg,
W.Hofbauer,
G.Lassmann,
P.Nordlund,
and
F.Lendzian
(2003).
Displacement of the tyrosyl radical cofactor in ribonucleotide reductase obtained by single-crystal high-field EPR and 1.4-A x-ray data.
|
| |
Proc Natl Acad Sci U S A,
100,
3209-3214.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
K.R.Strand,
S.Karlsen,
and
K.K.Andersson
(2002).
Cobalt substitution of mouse R2 ribonucleotide reductase as a model for the reactive diferrous state Spectroscopic and structural evidence for a ferromagnetically coupled dinuclear cobalt cluster.
|
| |
J Biol Chem,
277,
34229-34238.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

