 |
PDBsum entry 1jed

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|

|

|

|

|

|
 _CD
×11 _CD
×11
|
 |

|

|

|

|

|

|

|
 _CA
×6 _CA
×6
|
 |

|

|

|

|

|

|

|
 _NA
×12 _NA
×12
|
 |

|

|

|

|

|

|

|
 _MG
×4 _MG
×4
|
 |

|

|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.4
- sulfate adenylyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
sulfate + ATP + H+ = adenosine 5'-phosphosulfate + diphosphate
|
 |
 |
 |
 |
 |
sulfate
Bound ligand (Het Group name = )
matches with 87.10% similarity
|
+
|
 ATP
ATP
|
+
|
H(+)
|
=
|
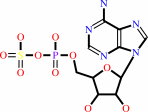 adenosine 5'-phosphosulfate
adenosine 5'-phosphosulfate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
313:1117-1125
(2001)
|
|
PubMed id:
|
|
|
|

|
| |
|
The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
|
|
T.C.Ullrich,
R.Huber.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The ubiquitous enzyme ATP sulfurylase (ATPS) catalyzes the primary step of
intracellular sulfate activation, the formation of adenosine 5'-phosphosulfate
(APS). It has been shown that the enzyme catalyzes the generation of APS from
ATP and inorganic sulfate in vitro and in vivo, and that this reaction can be
inhibited by a number of simple molecules. Here, we present the crystal
structures of ATPS from the yeast Saccharomyces cerevisiae complexed with
compounds that have inhibitory effects on the catalytic reaction of ATPS.
Thiosulfate and ADP mimic the substrates sulfate and ATP in the active site, but
are non-reactive and thus competitive inhibitors of the sulfurylase reaction.
Chlorate is bound in a crevice between the active site and the intermediate
domain III of the complex structure. It forms hydrogen bonds to residues of both
domains and stabilizes a "closed" conformation, inhibiting the release
of the reaction products APS and PPi. These new observations are evidence for
the crucial role of the displacement mechanism for the catalysis by ATPS.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Stereo view of the thiosulfate complex structure
of ATPS, showing the active site in standard orientation. The
final 2F[o] -F[c] electron density map at 2.5 Å around the
thiosulfate molecule, the cadmium ion and surrounding water
molecules is contoured at 1.0s.
|
 |
Figure 2.
Figure 2. Electrostatic surface potential plot of the ATPS
protomer (positive, blue; negative, red), showing (a) the active
site of the apo enzyme liganded with a sulfate molecule in its
binding pocket. The active site displays an open conformation
with the empty pocket for the adenosyl moiety of the nucleotide
on the left-hand of the sulfate group. (b) The ADP inhibitor
complex shows the active site in a closed conformation after
substrate recognition and rigid body displacement with the
envelope-like lid shielding the adenosyl moiety of ADP. The
putative magnesium ion is coloured in green. (c) Stereo plot of
the ADP complex structure of ATPS with the active site in
standard orientation. The final 2F[o] -F[c] electron density map
at 2.95 Å around ADP and liganding water molecules is
contoured at 1.0s. The green density at the phosphate groups of
ADP was interpreted and built as a magnesium ion (grey).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2001,
313,
1117-1125)
copyright 2001.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Oki,
K.Kitajima,
and
C.Meno
(2010).
Dissecting the role of Fgf signaling during gastrulation and left-right axis formation in mouse embryos using chemical inhibitors.
|
| |
Dev Dyn,
239,
1768-1778.
|
 |
|
|
|
|
 |
S.C.Gay,
I.H.Segel,
and
A.J.Fisher
(2009).
Structure of the two-domain hexameric APS kinase from Thiobacillus denitrificans: structural basis for the absence of ATP sulfurylase activity.
|
| |
Acta Crystallogr D Biol Crystallogr,
65,
1021-1031.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
O.Y.Gavel,
A.V.Kladova,
S.A.Bursakov,
J.M.Dias,
S.Texeira,
V.L.Shnyrov,
J.J.Moura,
I.Moura,
M.J.Romão,
and
J.Trincão
(2008).
Purification, crystallization and preliminary X-ray diffraction analysis of adenosine triphosphate sulfurylase (ATPS) from the sulfate-reducing bacterium Desulfovibrio desulfuricans ATCC 27774.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
593-595.
|
 |
|
|
|
|
 |
B.D.Spiegelberg,
J.Dela Cruz,
T.H.Law,
and
J.D.York
(2005).
Alteration of lithium pharmacology through manipulation of phosphoadenosine phosphate metabolism.
|
| |
J Biol Chem,
280,
5400-5405.
|
 |
|
|
|
|
 |
E.Hanna,
K.F.Ng,
I.J.MacRae,
C.J.Bley,
A.J.Fisher,
and
I.H.Segel
(2004).
Kinetic and stability properties of Penicillium chrysogenum ATP sulfurylase missing the C-terminal regulatory domain.
|
| |
J Biol Chem,
279,
4415-4424.
|
 |
|
|
|
|
 |
Y.Taguchi,
J.Hoseki,
Y.Kakuta,
and
K.Fukuyama
(2003).
Overproduction, crystallization and preliminary X-ray diffraction analysis of probable ATP sulfurylase from Thermus thermophilus HB8.
|
| |
Acta Crystallogr D Biol Crystallogr,
59,
1645-1647.
|
 |
|
|
|
|
 |
I.J.MacRae,
I.H.Segel,
and
A.J.Fisher
(2002).
Allosteric inhibition via R-state destabilization in ATP sulfurylase from Penicillium chrysogenum.
|
| |
Nat Struct Biol,
9,
945-949.
|
 |
|
PDB code:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

